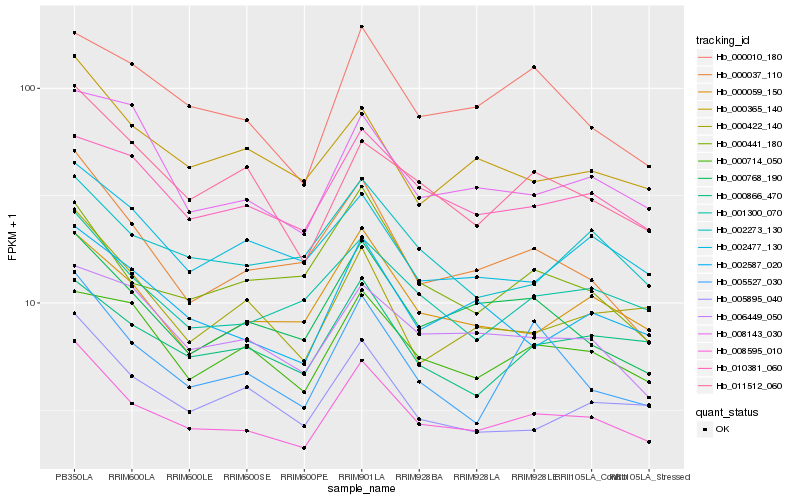
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008595_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06710, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001300_070 |
0.0787603159 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 3 |
Hb_000768_190 |
0.0827099032 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 4 |
Hb_000059_150 |
0.0846114985 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_002587_020 |
0.0848158502 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_010381_060 |
0.0901491967 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 7 |
Hb_000037_110 |
0.0914360766 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Populus euphratica] |
| 8 |
Hb_000866_470 |
0.092135687 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 9 |
Hb_005895_040 |
0.092788558 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 10 |
Hb_000365_140 |
0.0932671445 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 11 |
Hb_011512_060 |
0.0938954497 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000714_050 |
0.0939641004 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 13 |
Hb_006449_050 |
0.0950541524 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 14 |
Hb_000010_180 |
0.0951063021 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_002273_130 |
0.0951587911 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 16 |
Hb_002477_130 |
0.0970203679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000441_180 |
0.1005651035 |
- |
- |
Early endosome antigen, putative [Ricinus communis] |
| 18 |
Hb_000422_140 |
0.1009276292 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005527_030 |
0.1009433786 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g42630 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008143_030 |
0.1016476676 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |