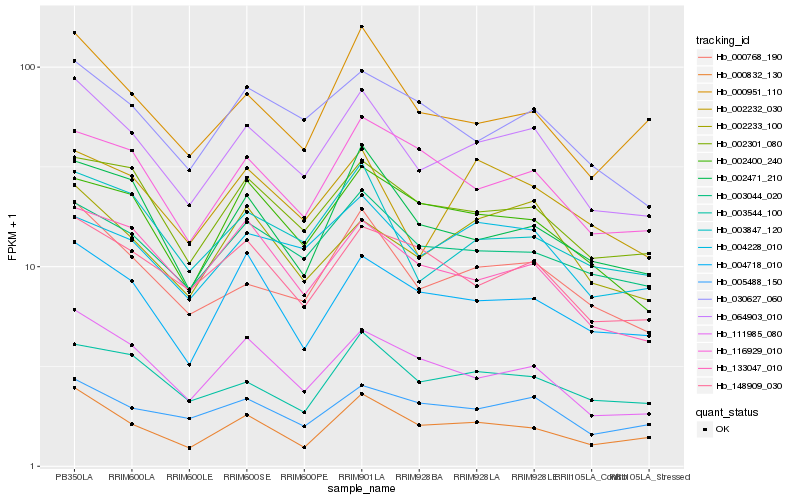
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064903_010 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 2 |
Hb_000768_190 |
0.0664858511 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 3 |
Hb_111985_080 |
0.0811792398 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 4 |
Hb_002301_080 |
0.0834351176 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003847_120 |
0.0877630115 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000832_130 |
0.0883593116 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g39620 [Jatropha curcas] |
| 7 |
Hb_003044_020 |
0.0883870656 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 8 |
Hb_030627_060 |
0.0908038646 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 9 |
Hb_005488_150 |
0.0916285362 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_133047_010 |
0.0931722414 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_002233_100 |
0.0933617601 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |
| 12 |
Hb_004228_010 |
0.0953681764 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA [Jatropha curcas] |
| 13 |
Hb_004718_010 |
0.0963510713 |
- |
- |
PREDICTED: uncharacterized protein LOC105638290 [Jatropha curcas] |
| 14 |
Hb_002471_210 |
0.0966935936 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 15 |
Hb_002232_030 |
0.0971261717 |
- |
- |
Pinin, putative [Ricinus communis] |
| 16 |
Hb_003544_100 |
0.0974241319 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |
| 17 |
Hb_000951_110 |
0.0977754004 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 18 |
Hb_148909_030 |
0.097914146 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002400_240 |
0.0992597038 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 20 |
Hb_116929_010 |
0.099351073 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |