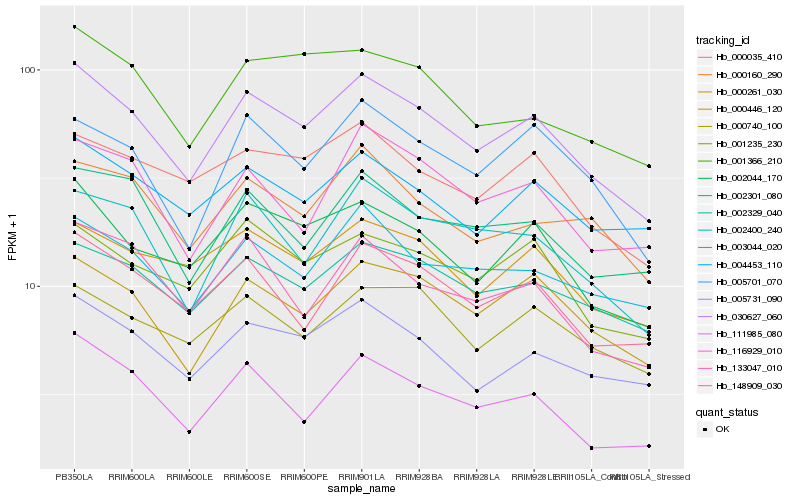
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030627_060 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 2 |
Hb_002044_170 |
0.0621664691 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 3 |
Hb_000446_120 |
0.0630761251 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 4 |
Hb_005731_090 |
0.0684243723 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 5 |
Hb_002329_040 |
0.0695112179 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 6 |
Hb_148909_030 |
0.0723656303 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001235_230 |
0.0727929508 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 8 |
Hb_002400_240 |
0.0745247451 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 9 |
Hb_003044_020 |
0.0745834793 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 10 |
Hb_005701_070 |
0.0747245268 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 11 |
Hb_000261_030 |
0.0759466519 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 12 |
Hb_000035_410 |
0.0772493374 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 13 |
Hb_111985_080 |
0.0775934306 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 14 |
Hb_002301_080 |
0.0778588501 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000160_290 |
0.0787480557 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_004453_110 |
0.0792304285 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000740_100 |
0.079285152 |
- |
- |
calpain, putative [Ricinus communis] |
| 18 |
Hb_001366_210 |
0.0810752766 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 19 |
Hb_116929_010 |
0.0819089369 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 20 |
Hb_133047_010 |
0.0820004054 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |