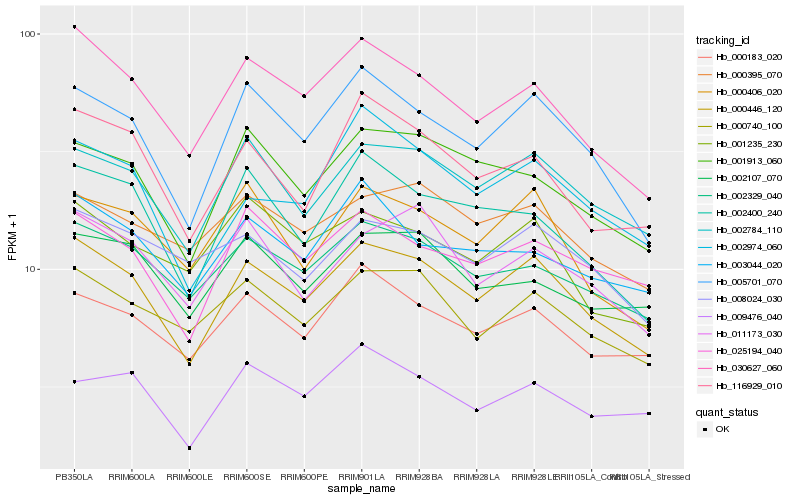
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000446_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 2 |
Hb_005701_070 |
0.0531952519 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 3 |
Hb_002784_110 |
0.0578463765 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 4 |
Hb_030627_060 |
0.0630761251 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 5 |
Hb_002329_040 |
0.0723343101 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 6 |
Hb_000740_100 |
0.0731729634 |
- |
- |
calpain, putative [Ricinus communis] |
| 7 |
Hb_025194_040 |
0.0738843264 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000183_020 |
0.0754348509 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001913_060 |
0.0755435186 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 10 |
Hb_116929_010 |
0.0775154653 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 11 |
Hb_000406_020 |
0.0794788584 |
- |
- |
- |
| 12 |
Hb_002400_240 |
0.0818086309 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_002974_060 |
0.0822887644 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 14 |
Hb_011173_030 |
0.0823827097 |
- |
- |
pumilio, putative [Ricinus communis] |
| 15 |
Hb_000395_070 |
0.0830454624 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003044_020 |
0.0869492063 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 17 |
Hb_008024_030 |
0.0876597995 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 18 |
Hb_002107_070 |
0.0881096949 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 19 |
Hb_001235_230 |
0.0886900902 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 20 |
Hb_009476_040 |
0.0887700889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |