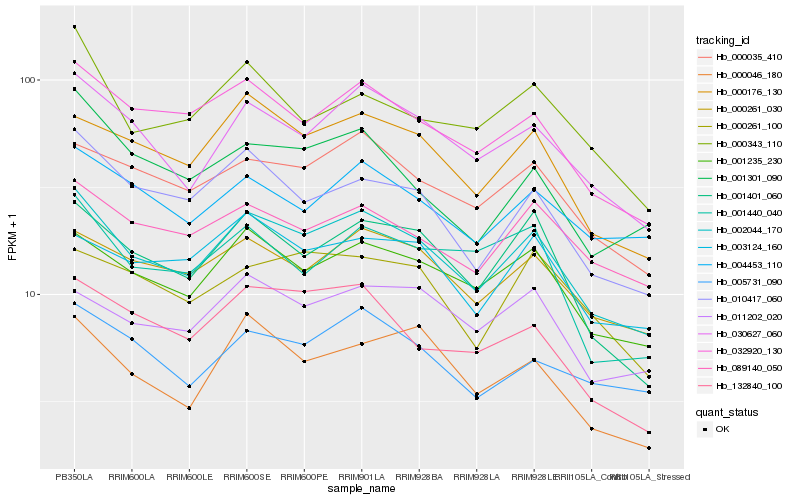
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002044_170 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 2 |
Hb_001235_230 |
0.0595302408 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 3 |
Hb_030627_060 |
0.0621664691 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 4 |
Hb_032920_130 |
0.0653182797 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 5 |
Hb_000261_030 |
0.0690718955 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 6 |
Hb_000176_130 |
0.0698121978 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 7 |
Hb_000035_410 |
0.0716705156 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 8 |
Hb_001401_060 |
0.0730909042 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 9 |
Hb_010417_060 |
0.0734127997 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 10 |
Hb_089140_050 |
0.0796500268 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 11 |
Hb_005731_090 |
0.0821376217 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 12 |
Hb_011202_020 |
0.0849790655 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003124_160 |
0.0854728112 |
- |
- |
dynamin, putative [Ricinus communis] |
| 14 |
Hb_004453_110 |
0.0859327534 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 15 |
Hb_132840_100 |
0.0890202553 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_000343_110 |
0.0898968213 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 17 |
Hb_001301_090 |
0.0902397823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000046_180 |
0.0902735173 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 19 |
Hb_000261_100 |
0.0918060035 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001440_040 |
0.0928424037 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |