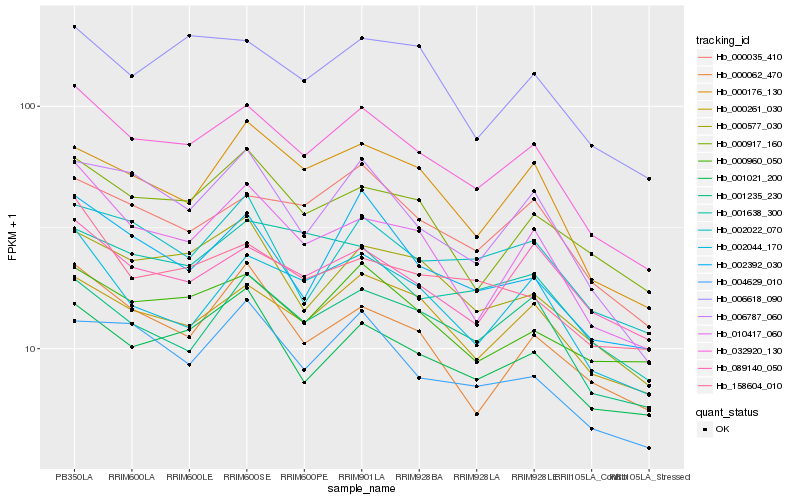
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032920_130 |
0.0 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 2 |
Hb_010417_060 |
0.0619038231 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 3 |
Hb_000261_030 |
0.0629722646 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 4 |
Hb_002044_170 |
0.0653182797 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 5 |
Hb_000035_410 |
0.0661506301 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 6 |
Hb_006787_060 |
0.068357284 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001235_230 |
0.070999332 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 8 |
Hb_000577_030 |
0.0713477214 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 9 |
Hb_000176_130 |
0.0728631083 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 10 |
Hb_002392_030 |
0.073127941 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 11 |
Hb_001021_200 |
0.0731519235 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 12 |
Hb_004629_010 |
0.0742894465 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 13 |
Hb_000917_160 |
0.0756759564 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006618_090 |
0.0757784009 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 15 |
Hb_000960_050 |
0.0775039231 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 16 |
Hb_158604_010 |
0.0785495422 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 17 |
Hb_089140_050 |
0.0790721058 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 18 |
Hb_000062_470 |
0.0809141094 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 19 |
Hb_001638_300 |
0.0827076759 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_002022_070 |
0.0836215849 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |