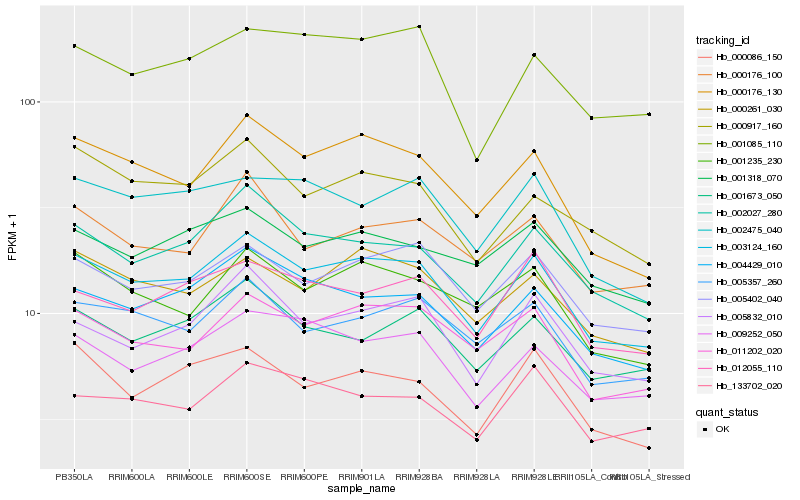
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_160 |
0.0 |
- |
- |
dynamin, putative [Ricinus communis] |
| 2 |
Hb_000176_130 |
0.0544202679 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 3 |
Hb_004429_010 |
0.0549270778 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_002027_280 |
0.0563009269 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 5 |
Hb_011202_020 |
0.0587749544 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005402_040 |
0.0590417149 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 7 |
Hb_000261_030 |
0.0626739908 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 8 |
Hb_005357_260 |
0.0632879023 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 9 |
Hb_005832_010 |
0.064368675 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009252_050 |
0.0654405622 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_001235_230 |
0.0673697827 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_001673_050 |
0.0676534183 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002475_040 |
0.0692899751 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_012055_110 |
0.0705185142 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 15 |
Hb_000176_100 |
0.0717757404 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 16 |
Hb_001318_070 |
0.0721013554 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000917_160 |
0.0723161143 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000086_150 |
0.0723192056 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 19 |
Hb_133702_020 |
0.072338452 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001085_110 |
0.0724460652 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |