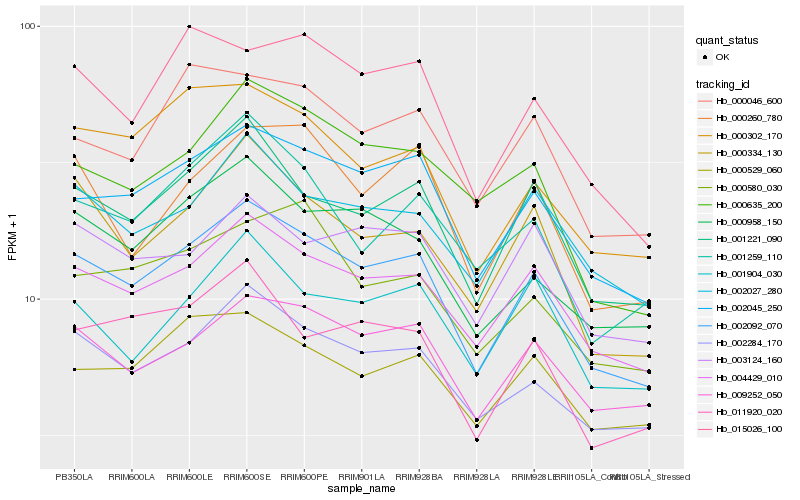
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002092_070 |
0.0 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 2 |
Hb_002045_250 |
0.0477133454 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 3 |
Hb_004429_010 |
0.0532140253 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_002284_170 |
0.056820367 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 5 |
Hb_001221_090 |
0.0570834864 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009252_050 |
0.067664529 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000958_150 |
0.0683162223 |
- |
- |
Actin, putative [Ricinus communis] |
| 8 |
Hb_000334_130 |
0.0731846563 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 9 |
Hb_000302_170 |
0.0741005228 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 10 |
Hb_002027_280 |
0.0750184896 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 11 |
Hb_000046_600 |
0.0754071948 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 12 |
Hb_011920_020 |
0.0761279619 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 13 |
Hb_015026_100 |
0.0779249275 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 14 |
Hb_000529_060 |
0.0786322752 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000635_200 |
0.0804036104 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 16 |
Hb_000260_780 |
0.0810709829 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |
| 17 |
Hb_003124_160 |
0.0814667296 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_001904_030 |
0.0823160786 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 19 |
Hb_000580_030 |
0.0826303459 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 20 |
Hb_001259_110 |
0.0834106283 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |