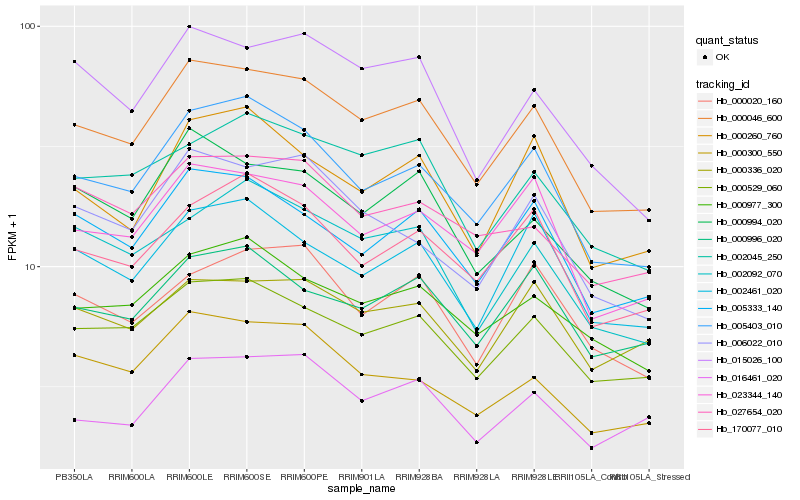
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_600 |
0.0 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 2 |
Hb_005403_010 |
0.0526356054 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 3 |
Hb_000529_060 |
0.0528366469 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 4 |
Hb_023344_140 |
0.0591886147 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 5 |
Hb_005333_140 |
0.0606584489 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000994_020 |
0.0662735827 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000977_300 |
0.0675782244 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 8 |
Hb_002045_250 |
0.068900137 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 9 |
Hb_170077_010 |
0.0694684934 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_015026_100 |
0.0701600312 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 11 |
Hb_000260_760 |
0.0716933544 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 12 |
Hb_000996_020 |
0.0729117189 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 13 |
Hb_000300_550 |
0.0734393236 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_002461_020 |
0.0735520846 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 15 |
Hb_027654_020 |
0.0744126053 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 16 |
Hb_002092_070 |
0.0754071948 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 17 |
Hb_006022_010 |
0.0764415166 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 18 |
Hb_016461_020 |
0.0773583692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000336_020 |
0.0785336455 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 20 |
Hb_000020_160 |
0.0796926902 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |