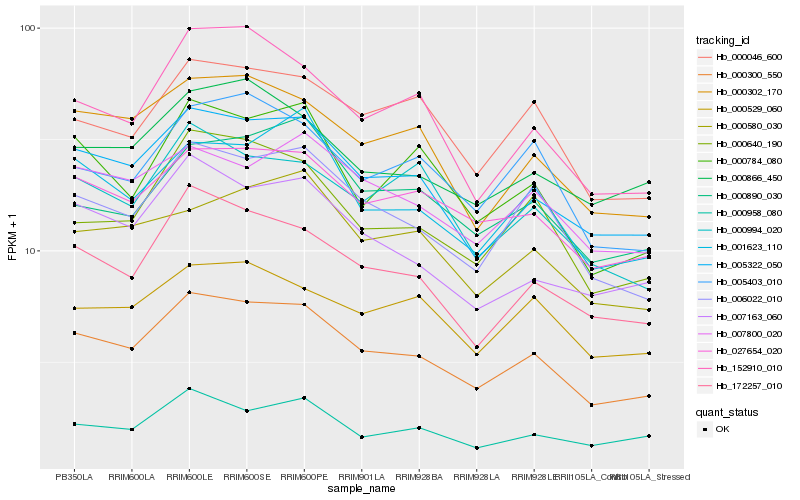
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_550 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 2 |
Hb_005322_050 |
0.0451916079 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 3 |
Hb_006022_010 |
0.0596252829 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 4 |
Hb_027654_020 |
0.0602101176 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 5 |
Hb_172257_010 |
0.0621744396 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 6 |
Hb_000302_170 |
0.0634147302 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 7 |
Hb_000640_190 |
0.0696963684 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000046_600 |
0.0734393236 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 9 |
Hb_007163_060 |
0.0744827663 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 10 |
Hb_001623_110 |
0.076415181 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_152910_010 |
0.0765525843 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 12 |
Hb_005403_010 |
0.0769325509 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 13 |
Hb_007800_020 |
0.077293267 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000866_450 |
0.0784673036 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000958_080 |
0.0799002248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000784_080 |
0.0810174002 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 17 |
Hb_000890_030 |
0.0814709016 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 18 |
Hb_000580_030 |
0.0821253485 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 19 |
Hb_000994_020 |
0.082750127 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000529_060 |
0.0828755194 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |