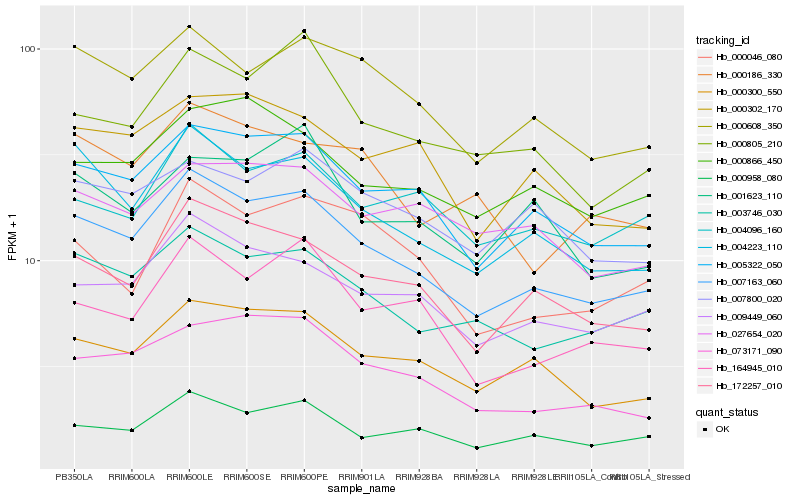
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007163_060 |
0.0 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 2 |
Hb_005322_050 |
0.0590651345 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 3 |
Hb_172257_010 |
0.0670115656 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 4 |
Hb_004223_110 |
0.0675392299 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 5 |
Hb_003746_030 |
0.0734659969 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 6 |
Hb_000300_550 |
0.0744827663 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 7 |
Hb_009449_060 |
0.0765400855 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 8 |
Hb_000608_350 |
0.0817200212 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 9 |
Hb_000958_080 |
0.0828372462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_073171_090 |
0.0869855095 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 11 |
Hb_164945_010 |
0.0928295325 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 12 |
Hb_000805_210 |
0.0949323956 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 13 |
Hb_000302_170 |
0.0961685512 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 14 |
Hb_000866_450 |
0.0996578272 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_004096_160 |
0.099711038 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 16 |
Hb_000046_080 |
0.1031325117 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |
| 17 |
Hb_007800_020 |
0.103134526 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000186_330 |
0.1041621534 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 19 |
Hb_027654_020 |
0.1081866888 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 20 |
Hb_001623_110 |
0.1090032806 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |