| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
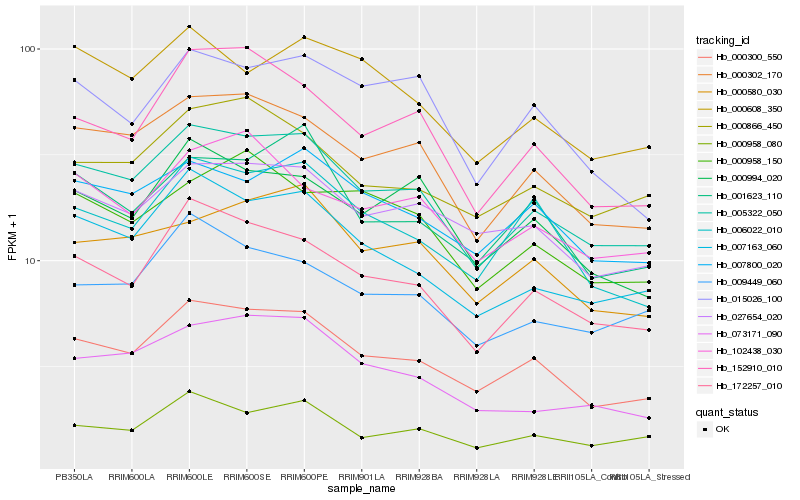
Hb_005322_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 2 |
Hb_000300_550 |
0.0451916079 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 3 |
Hb_000302_170 |
0.0492064697 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 4 |
Hb_172257_010 |
0.0576192881 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 5 |
Hb_007163_060 |
0.0590651345 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 6 |
Hb_000958_080 |
0.0720672825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_027654_020 |
0.0749266188 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 8 |
Hb_073171_090 |
0.0776746621 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 9 |
Hb_007800_020 |
0.0785664366 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000580_030 |
0.0789381752 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 11 |
Hb_009449_060 |
0.079204294 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 12 |
Hb_152910_010 |
0.0807105065 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 13 |
Hb_000866_450 |
0.0822349461 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_000994_020 |
0.084161839 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_001623_110 |
0.0845566188 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_006022_010 |
0.0852785192 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 17 |
Hb_000608_350 |
0.0855744245 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 18 |
Hb_102438_030 |
0.0879911798 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000958_150 |
0.0910878257 |
- |
- |
Actin, putative [Ricinus communis] |
| 20 |
Hb_015026_100 |
0.0910981308 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |