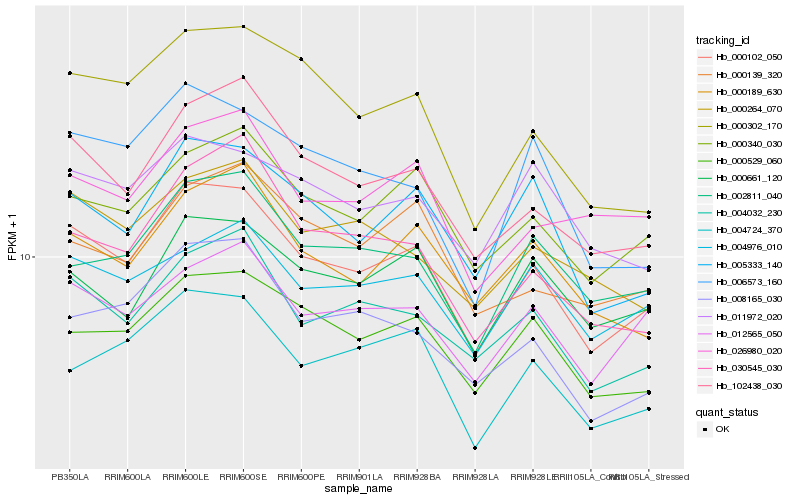
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000102_050 |
0.0 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_102438_030 |
0.0578947882 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004032_230 |
0.0642304104 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 4 |
Hb_000340_030 |
0.0671496036 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 5 |
Hb_000529_060 |
0.0680668847 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005333_140 |
0.0706505205 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000302_170 |
0.072395918 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 8 |
Hb_012565_050 |
0.0742005201 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 9 |
Hb_008165_030 |
0.0747523285 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 10 |
Hb_004724_370 |
0.0748531079 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 11 |
Hb_000189_630 |
0.0756693358 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 12 |
Hb_002811_040 |
0.0786171184 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 13 |
Hb_004976_010 |
0.0794386952 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 14 |
Hb_000264_070 |
0.0794440428 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 15 |
Hb_000661_120 |
0.0800041663 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 16 |
Hb_006573_160 |
0.0810657316 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 17 |
Hb_030545_030 |
0.0814368678 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 18 |
Hb_026980_020 |
0.0840564683 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 19 |
Hb_000139_320 |
0.0849316349 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 20 |
Hb_011972_020 |
0.085130016 |
- |
- |
protein binding protein, putative [Ricinus communis] |