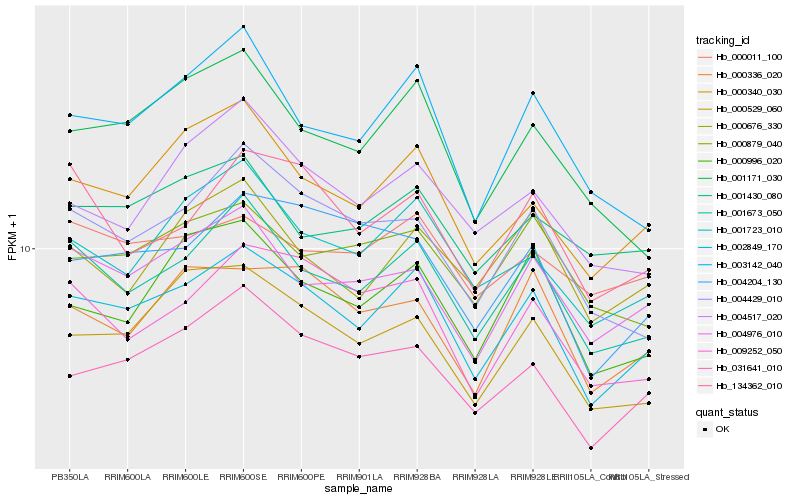
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001673_050 |
0.0492266023 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000340_030 |
0.0586899129 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 4 |
Hb_031641_010 |
0.0604679668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001723_010 |
0.0636773363 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 6 |
Hb_000529_060 |
0.0656640509 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004976_010 |
0.0659502544 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 8 |
Hb_000336_020 |
0.0687234621 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 9 |
Hb_000996_020 |
0.068864623 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 10 |
Hb_009252_050 |
0.0711885783 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_003142_040 |
0.0714318223 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 12 |
Hb_004429_010 |
0.0729661828 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 13 |
Hb_004204_130 |
0.073353812 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 14 |
Hb_134362_010 |
0.0739843302 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 15 |
Hb_000879_040 |
0.0752356331 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 16 |
Hb_001171_030 |
0.075493859 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 17 |
Hb_000676_330 |
0.0765078368 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_004517_020 |
0.077822063 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 19 |
Hb_000011_100 |
0.0785904688 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 20 |
Hb_001430_080 |
0.0788688971 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |