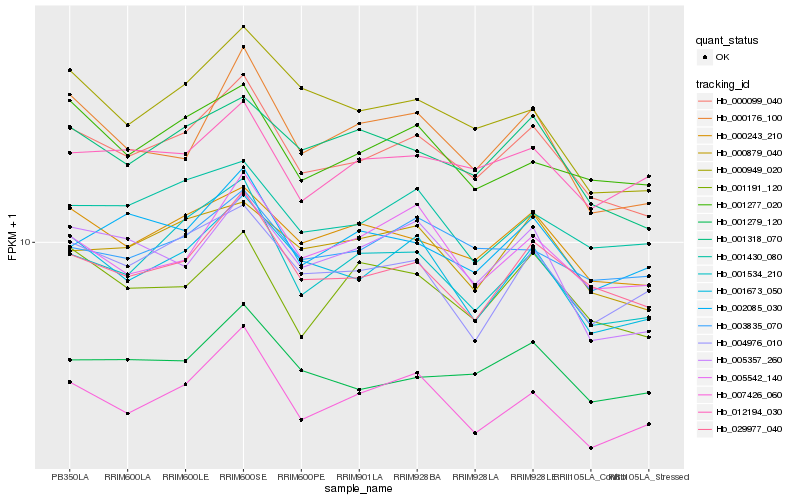
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_040 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_029977_040 |
0.0467002906 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 3 |
Hb_000243_210 |
0.0511813778 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_001673_050 |
0.0537539479 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001318_070 |
0.0571178529 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001279_120 |
0.0583353732 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 7 |
Hb_001534_210 |
0.0589994839 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000879_040 |
0.0617853738 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 9 |
Hb_001430_080 |
0.0630160159 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 10 |
Hb_000176_100 |
0.0630421809 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 11 |
Hb_007426_060 |
0.0634851419 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_001277_020 |
0.0644932476 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 13 |
Hb_003835_070 |
0.0655101018 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 14 |
Hb_012194_030 |
0.0663713931 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000949_020 |
0.0663950366 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 16 |
Hb_001191_120 |
0.0664476028 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005542_140 |
0.0665184271 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 18 |
Hb_002085_030 |
0.0670933768 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004976_010 |
0.0675253701 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 20 |
Hb_005357_260 |
0.0685062526 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |