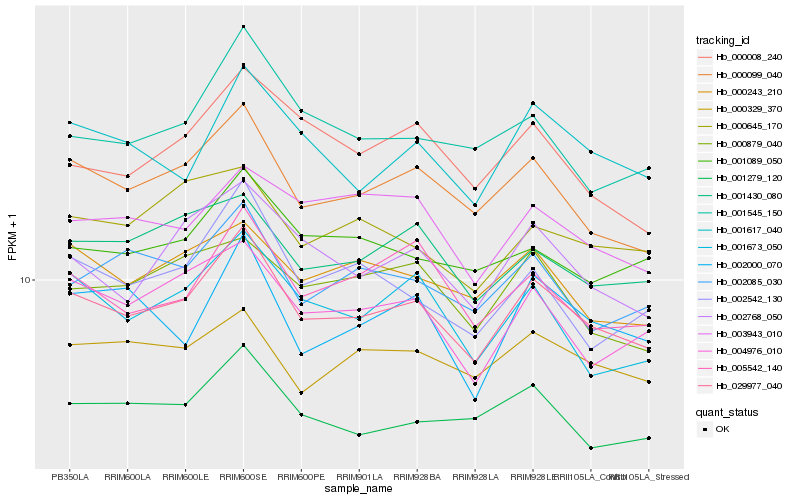
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029977_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000099_040 |
0.0467002906 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_002768_050 |
0.0630461186 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 4 |
Hb_001617_040 |
0.0637893648 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_004976_010 |
0.0638889677 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 6 |
Hb_001279_120 |
0.0639302599 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 7 |
Hb_005542_140 |
0.0664196171 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 8 |
Hb_001673_050 |
0.0666701409 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001430_080 |
0.0674855338 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 10 |
Hb_000243_210 |
0.0677656622 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_002085_030 |
0.067986134 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000329_370 |
0.069652869 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 13 |
Hb_001089_050 |
0.0700122331 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 14 |
Hb_002542_130 |
0.0705856651 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 15 |
Hb_001545_150 |
0.0712772406 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 16 |
Hb_000879_040 |
0.0732563471 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 17 |
Hb_002000_070 |
0.0734753013 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 18 |
Hb_000008_240 |
0.0738311362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000645_170 |
0.0738680964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003943_010 |
0.0741169187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |