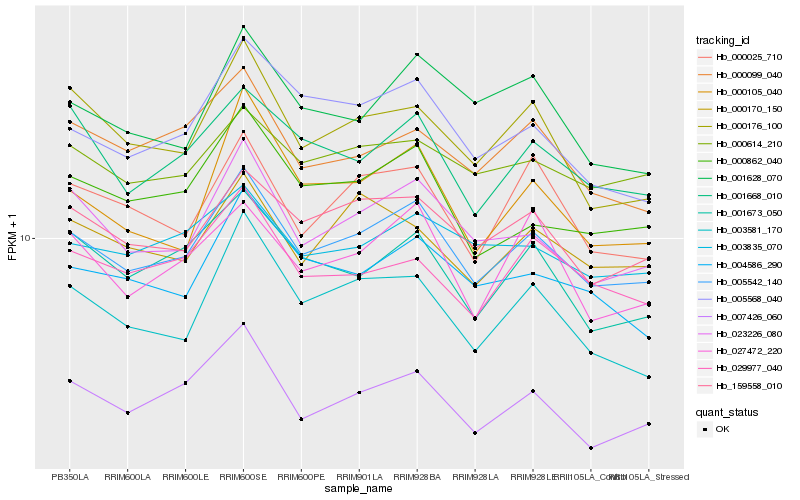
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_140 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 2 |
Hb_023226_080 |
0.0503647925 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 3 |
Hb_159558_010 |
0.0509980571 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 4 |
Hb_007426_060 |
0.0587571999 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000862_040 |
0.0595311473 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 6 |
Hb_005568_040 |
0.0597516877 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001668_010 |
0.0617038134 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 8 |
Hb_000176_100 |
0.0624354369 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 9 |
Hb_004586_290 |
0.0625122893 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 10 |
Hb_000614_210 |
0.0637006551 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001673_050 |
0.0647005185 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000025_710 |
0.0650181566 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 13 |
Hb_003835_070 |
0.0652356448 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003581_170 |
0.0656034719 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_029977_040 |
0.0664196171 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 16 |
Hb_000099_040 |
0.0665184271 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000105_040 |
0.0666634702 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 18 |
Hb_001628_070 |
0.0667138923 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_027472_220 |
0.0675182713 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000170_150 |
0.0683619688 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |