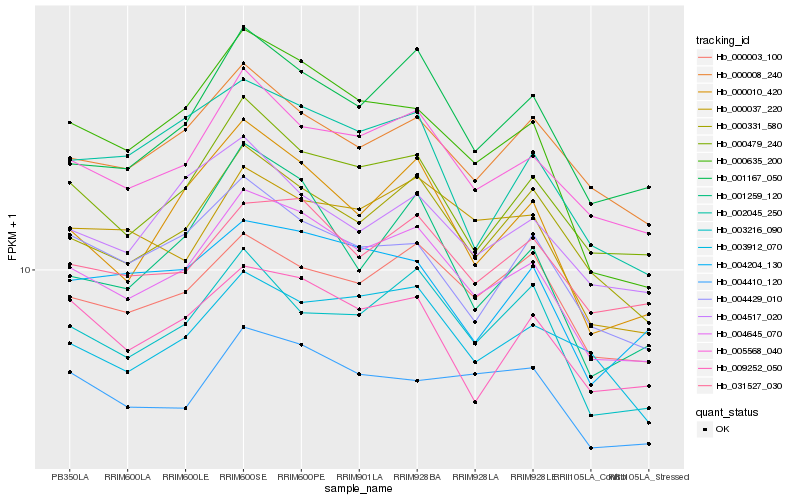
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004645_070 |
0.0 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 2 |
Hb_005568_040 |
0.0512381923 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000479_240 |
0.0583726197 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000010_420 |
0.062083794 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 5 |
Hb_000331_580 |
0.0621734391 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 6 |
Hb_000635_200 |
0.0627386739 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 7 |
Hb_001167_050 |
0.066404521 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 8 |
Hb_031527_030 |
0.0689515623 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_009252_050 |
0.0705995408 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000003_100 |
0.0716507378 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 11 |
Hb_003216_090 |
0.0718354842 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000008_240 |
0.0719119179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000037_220 |
0.0729505744 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 14 |
Hb_004429_010 |
0.0735223101 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_002045_250 |
0.0756066081 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004410_120 |
0.0756523256 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004204_130 |
0.0776611738 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 18 |
Hb_003912_070 |
0.0781006641 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 19 |
Hb_004517_020 |
0.0794087074 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 20 |
Hb_001259_120 |
0.0799626285 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |