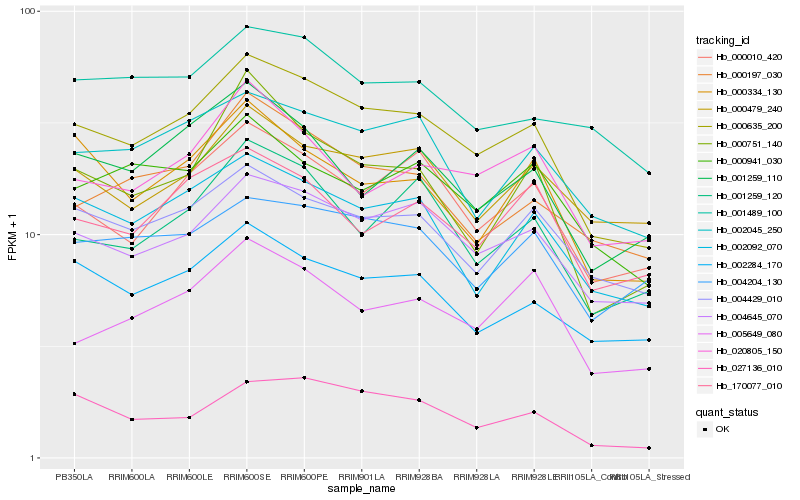
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_200 |
0.0 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 2 |
Hb_004645_070 |
0.0627386739 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 3 |
Hb_002092_070 |
0.0804036104 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 4 |
Hb_000010_420 |
0.0816630134 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 5 |
Hb_002045_250 |
0.0834803358 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 6 |
Hb_004429_010 |
0.0840926995 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 7 |
Hb_002284_170 |
0.0876180859 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 8 |
Hb_001259_120 |
0.0898025512 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 9 |
Hb_000197_030 |
0.0920441183 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 10 |
Hb_000941_030 |
0.0923902302 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 11 |
Hb_027136_010 |
0.0952601888 |
- |
- |
hypothetical protein L484_010426 [Morus notabilis] |
| 12 |
Hb_000334_130 |
0.0959451268 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 13 |
Hb_001259_110 |
0.096036795 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 14 |
Hb_170077_010 |
0.0971592199 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 15 |
Hb_020805_150 |
0.0972841651 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_000751_140 |
0.0974460214 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 17 |
Hb_005649_080 |
0.0975270417 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 18 |
Hb_000479_240 |
0.0975563117 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001489_100 |
0.0978981859 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 20 |
Hb_004204_130 |
0.0983325965 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |