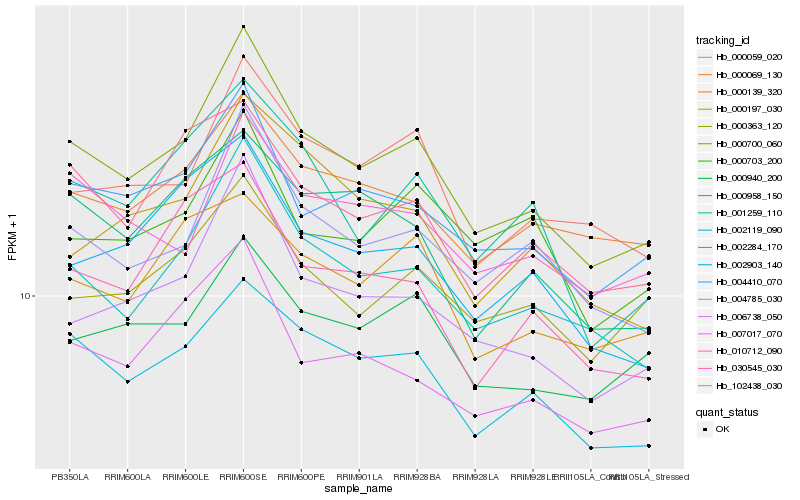
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 2 |
Hb_002119_090 |
0.0525254604 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 3 |
Hb_004785_030 |
0.0621816656 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 4 |
Hb_006738_050 |
0.0622698027 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 5 |
Hb_002284_170 |
0.0632823097 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 6 |
Hb_004410_070 |
0.0681130735 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 7 |
Hb_000059_020 |
0.0740511769 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 8 |
Hb_007017_070 |
0.0774000617 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001259_110 |
0.0792380157 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 10 |
Hb_000069_130 |
0.0798552792 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 11 |
Hb_102438_030 |
0.0824819715 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_010712_090 |
0.083082075 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 13 |
Hb_000940_200 |
0.0831511581 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 14 |
Hb_000363_120 |
0.0845252493 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 15 |
Hb_002903_140 |
0.0852327695 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 16 |
Hb_000958_150 |
0.0859414679 |
- |
- |
Actin, putative [Ricinus communis] |
| 17 |
Hb_000197_030 |
0.0880189821 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 18 |
Hb_000139_320 |
0.0889731961 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 19 |
Hb_030545_030 |
0.0897233244 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 20 |
Hb_000703_200 |
0.0904128596 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |