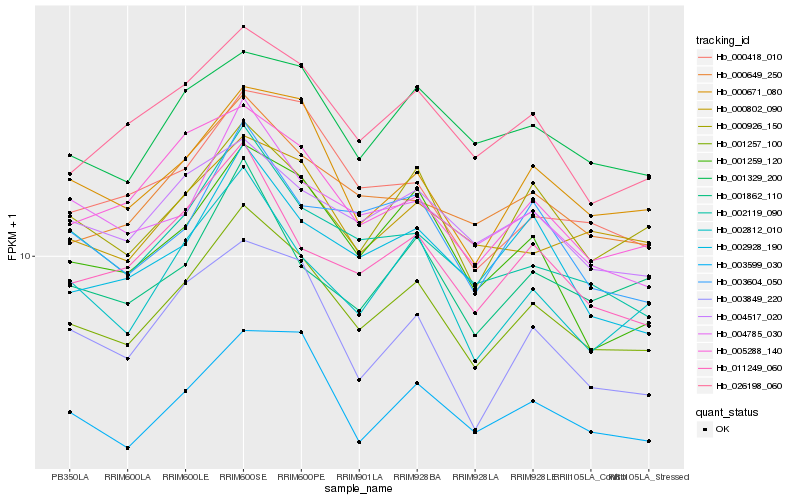
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001257_100 |
0.0 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 2 |
Hb_000671_080 |
0.0691580205 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 3 |
Hb_000926_150 |
0.0716727263 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_026198_060 |
0.0746508863 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 5 |
Hb_002812_010 |
0.0746867857 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000649_250 |
0.0754203923 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 7 |
Hb_002119_090 |
0.0775594901 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 8 |
Hb_005288_140 |
0.0797597294 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 9 |
Hb_001259_120 |
0.0805905539 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 10 |
Hb_011249_060 |
0.0814154919 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 11 |
Hb_001329_200 |
0.0836295169 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 12 |
Hb_003599_030 |
0.0838011052 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_004785_030 |
0.0839830534 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 14 |
Hb_002928_190 |
0.0842803056 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 15 |
Hb_003604_050 |
0.0844297918 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_004517_020 |
0.0850628043 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 17 |
Hb_000418_010 |
0.0851633357 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 18 |
Hb_003849_220 |
0.0866144181 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 19 |
Hb_000802_090 |
0.086997043 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_001862_110 |
0.0876919394 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |