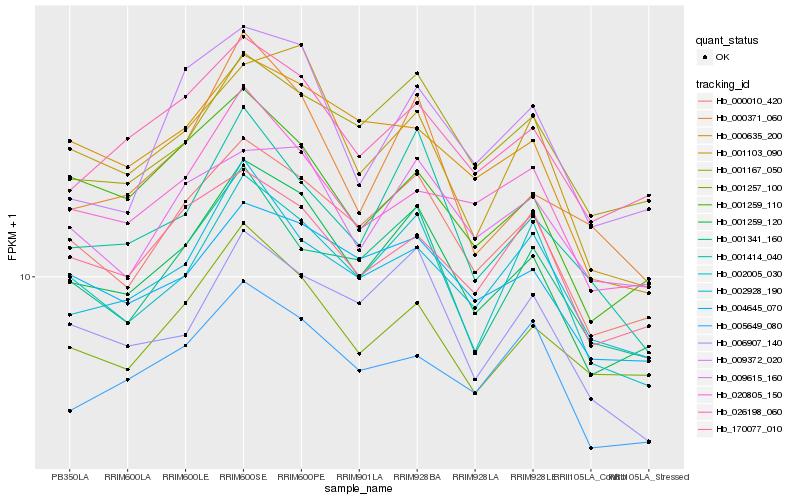
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001259_120 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 2 |
Hb_000010_420 |
0.0506738304 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 3 |
Hb_026198_060 |
0.0756533968 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 4 |
Hb_004645_070 |
0.0799626285 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 5 |
Hb_001257_100 |
0.0805905539 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_002005_030 |
0.080708033 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 7 |
Hb_001167_050 |
0.0828533303 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 8 |
Hb_006907_140 |
0.0877466322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001259_110 |
0.0877794698 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 10 |
Hb_001103_090 |
0.088314671 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 11 |
Hb_001341_160 |
0.088703249 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001414_040 |
0.089368909 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000635_200 |
0.0898025512 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 14 |
Hb_009615_160 |
0.0898663186 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 15 |
Hb_002928_190 |
0.0919198589 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 16 |
Hb_000371_060 |
0.0920023102 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 17 |
Hb_170077_010 |
0.0938261179 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_020805_150 |
0.0948531131 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_005649_080 |
0.0954489819 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 20 |
Hb_009372_020 |
0.0971100279 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |