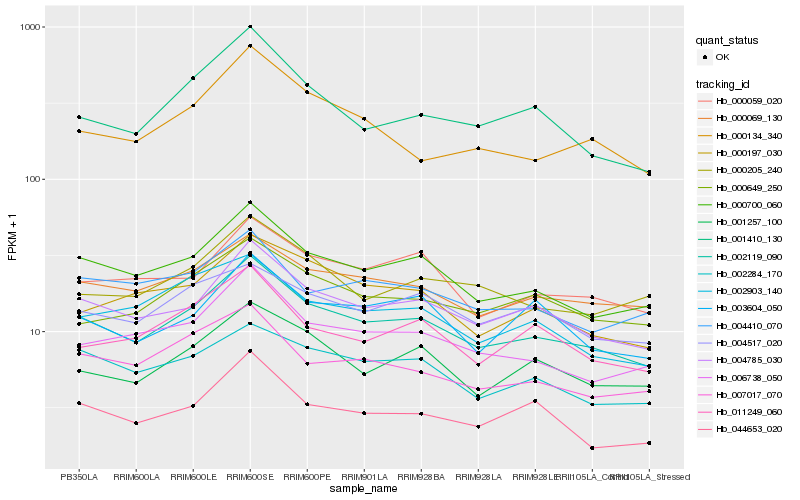
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002119_090 |
0.0 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 2 |
Hb_004785_030 |
0.0514058851 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 3 |
Hb_000700_060 |
0.0525254604 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 4 |
Hb_000069_130 |
0.075055954 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006738_050 |
0.0752897336 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 6 |
Hb_001257_100 |
0.0775594901 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 7 |
Hb_002284_170 |
0.0814382724 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 8 |
Hb_007017_070 |
0.0815487646 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000059_020 |
0.0820516469 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 10 |
Hb_001410_130 |
0.083393375 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 11 |
Hb_000649_250 |
0.0846589608 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 12 |
Hb_011249_060 |
0.0849768128 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 13 |
Hb_000197_030 |
0.0866955339 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 14 |
Hb_044653_020 |
0.0873485686 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 15 |
Hb_000205_240 |
0.0876351038 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_003604_050 |
0.0880945197 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002903_140 |
0.0886116741 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 18 |
Hb_004517_020 |
0.0887123162 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 19 |
Hb_000134_340 |
0.090277581 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 20 |
Hb_004410_070 |
0.0905757796 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |