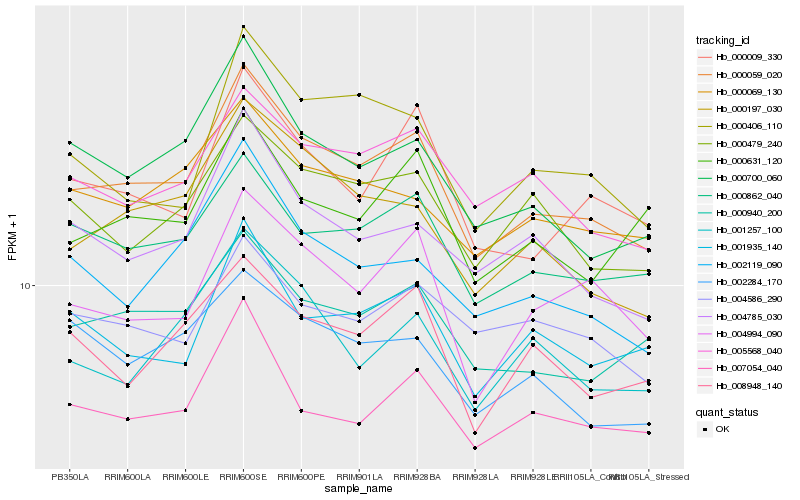
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_020 |
0.0 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 2 |
Hb_000862_040 |
0.0691949458 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 3 |
Hb_000940_200 |
0.0708684763 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 4 |
Hb_005568_040 |
0.0738908856 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000700_060 |
0.0740511769 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 6 |
Hb_000197_030 |
0.0770321923 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 7 |
Hb_000009_330 |
0.07758078 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_000479_240 |
0.077819756 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000069_130 |
0.0787706076 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004785_030 |
0.0788959667 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 11 |
Hb_004994_090 |
0.0790345403 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007054_040 |
0.0812044473 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 13 |
Hb_001935_140 |
0.0816415666 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 14 |
Hb_002119_090 |
0.0820516469 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 15 |
Hb_008948_140 |
0.0831549114 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 16 |
Hb_000406_110 |
0.0853380842 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 17 |
Hb_002284_170 |
0.0865466907 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 18 |
Hb_004586_290 |
0.0878766806 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 19 |
Hb_000631_120 |
0.0880394275 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 20 |
Hb_001257_100 |
0.088273189 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |