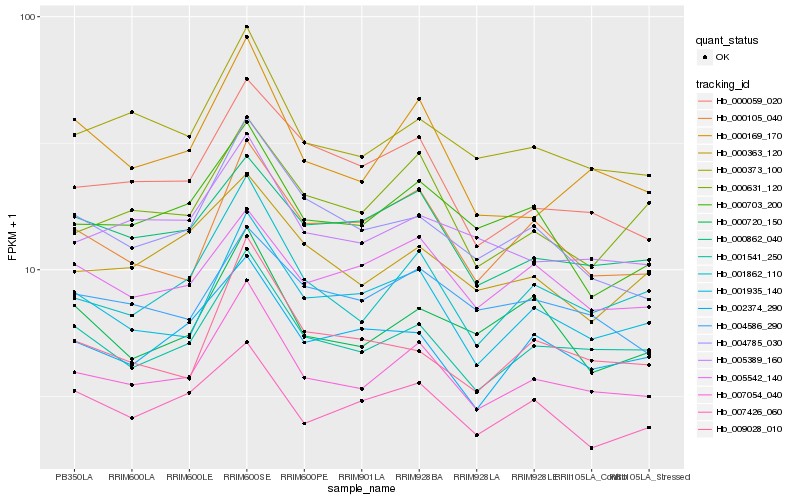
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007054_040 |
0.0 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 2 |
Hb_001541_250 |
0.0505996354 |
- |
- |
PREDICTED: integrator complex subunit 9 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_001862_110 |
0.0525821252 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_000373_100 |
0.0614495125 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 5 |
Hb_001935_140 |
0.0655680817 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 6 |
Hb_005389_160 |
0.0745499844 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 7 |
Hb_000169_170 |
0.0754944875 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 8 |
Hb_000862_040 |
0.0763360594 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 9 |
Hb_000105_040 |
0.0777609786 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 10 |
Hb_000059_020 |
0.0812044473 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 11 |
Hb_002374_290 |
0.0814390028 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000703_200 |
0.0814537325 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004785_030 |
0.0815126431 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 14 |
Hb_000631_120 |
0.0828649864 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 15 |
Hb_005542_140 |
0.0843333712 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 16 |
Hb_004586_290 |
0.0847949745 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 17 |
Hb_000720_150 |
0.0849505504 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 18 |
Hb_007426_060 |
0.0854416519 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_009028_010 |
0.0856351224 |
- |
- |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 20 |
Hb_000363_120 |
0.0873748725 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |