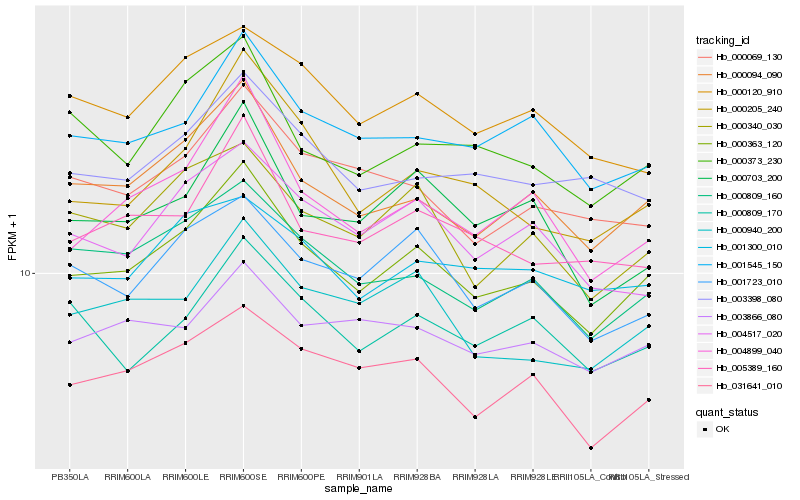
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_120 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 2 |
Hb_000094_090 |
0.0462273769 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000809_160 |
0.0604167356 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 4 |
Hb_031641_010 |
0.0649005164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000340_030 |
0.0650113536 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 6 |
Hb_004899_040 |
0.0652018597 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 7 |
Hb_001545_150 |
0.0662175618 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 8 |
Hb_001723_010 |
0.0690611214 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 9 |
Hb_000940_200 |
0.0691968068 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 10 |
Hb_005389_160 |
0.0704500147 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 11 |
Hb_000205_240 |
0.0717099572 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_004517_020 |
0.0726189682 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 13 |
Hb_000373_230 |
0.0733019348 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000069_130 |
0.0738507905 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000703_200 |
0.0741924943 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003398_080 |
0.075824385 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000120_910 |
0.07656563 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 18 |
Hb_000809_170 |
0.0770998365 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_001300_010 |
0.0776081355 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_003866_080 |
0.0792332855 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |