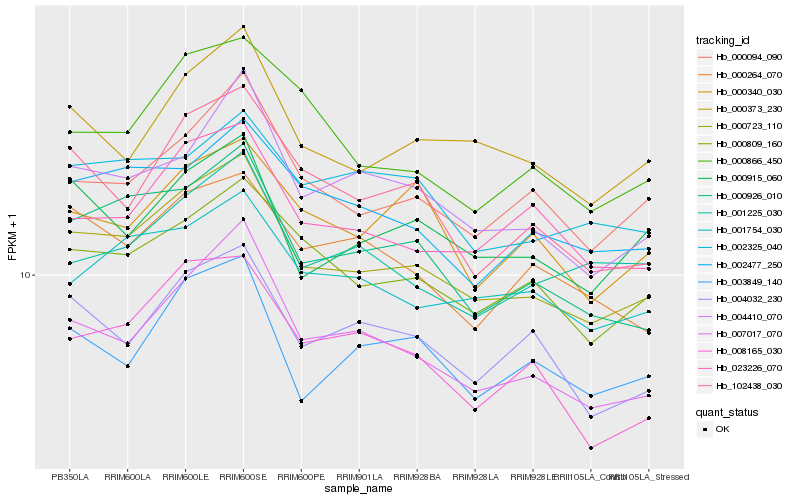
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_110 |
0.0 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 2 |
Hb_007017_070 |
0.0575548938 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000926_010 |
0.0585749431 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 4 |
Hb_000809_160 |
0.0609591825 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 5 |
Hb_004410_070 |
0.0624410241 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 6 |
Hb_000094_090 |
0.0625679613 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000373_230 |
0.0646780452 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 8 |
Hb_001754_030 |
0.0682573434 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 9 |
Hb_102438_030 |
0.0702706861 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_023226_070 |
0.0704259704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004032_230 |
0.0708570278 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 12 |
Hb_002477_250 |
0.072523755 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 13 |
Hb_000915_060 |
0.0728001524 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 14 |
Hb_003849_140 |
0.0738814561 |
- |
- |
PREDICTED: uncharacterized protein LOC105644697 [Jatropha curcas] |
| 15 |
Hb_000866_450 |
0.0757380206 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_008165_030 |
0.0782146514 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 17 |
Hb_000264_070 |
0.078421798 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 18 |
Hb_002325_040 |
0.0805720332 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001225_030 |
0.0811979725 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 20 |
Hb_000340_030 |
0.0815041483 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |