| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
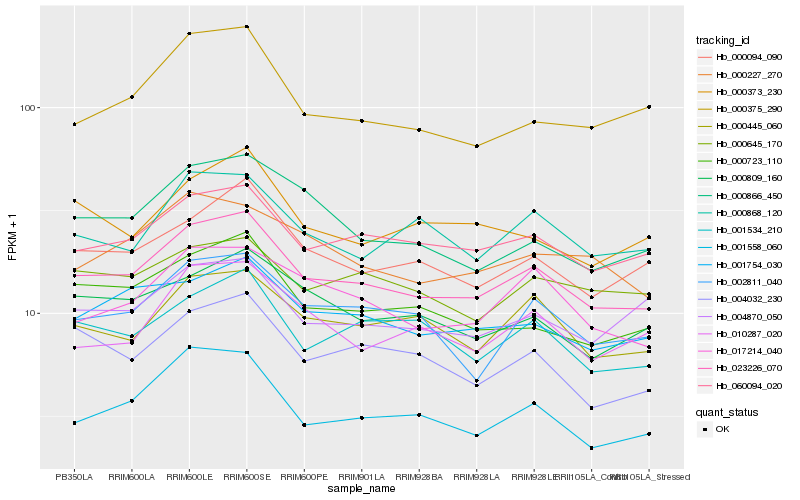
Hb_023226_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_060094_020 |
0.0549884493 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000094_090 |
0.0646463978 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001754_030 |
0.0651802458 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 5 |
Hb_000445_060 |
0.0655369268 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000723_110 |
0.0704259704 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 7 |
Hb_002811_040 |
0.0711852766 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 8 |
Hb_001534_210 |
0.0722331542 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000809_160 |
0.0722959557 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 10 |
Hb_017214_040 |
0.0724367828 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000868_120 |
0.0732594485 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 12 |
Hb_004870_050 |
0.0748098488 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 13 |
Hb_004032_230 |
0.0751742031 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 14 |
Hb_001558_060 |
0.0755402264 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000227_270 |
0.0769501804 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010287_020 |
0.077378197 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 17 |
Hb_000645_170 |
0.0777523968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000866_450 |
0.078928469 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000375_290 |
0.0789689365 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000373_230 |
0.0791551593 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |