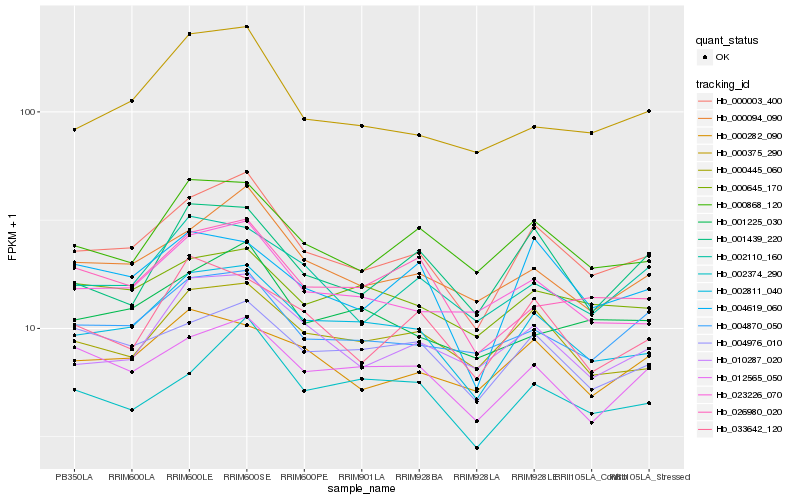
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_400 |
0.0 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_002811_040 |
0.0566028466 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 3 |
Hb_000094_090 |
0.0702792431 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000868_120 |
0.0777445617 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 5 |
Hb_023226_070 |
0.0805322958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010287_020 |
0.0814349607 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 7 |
Hb_000445_060 |
0.0816694626 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 8 |
Hb_004976_010 |
0.0836411666 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 9 |
Hb_002374_290 |
0.0842024911 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004870_050 |
0.0859148855 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 11 |
Hb_012565_050 |
0.0865051591 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 12 |
Hb_000375_290 |
0.0896372225 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_002110_160 |
0.0897804136 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001439_220 |
0.090008724 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 15 |
Hb_001225_030 |
0.0906326927 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 16 |
Hb_026980_020 |
0.0907702958 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 17 |
Hb_004619_060 |
0.0909520952 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 18 |
Hb_000282_090 |
0.092328755 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 19 |
Hb_033642_120 |
0.0925565937 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 20 |
Hb_000645_170 |
0.0928315213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |