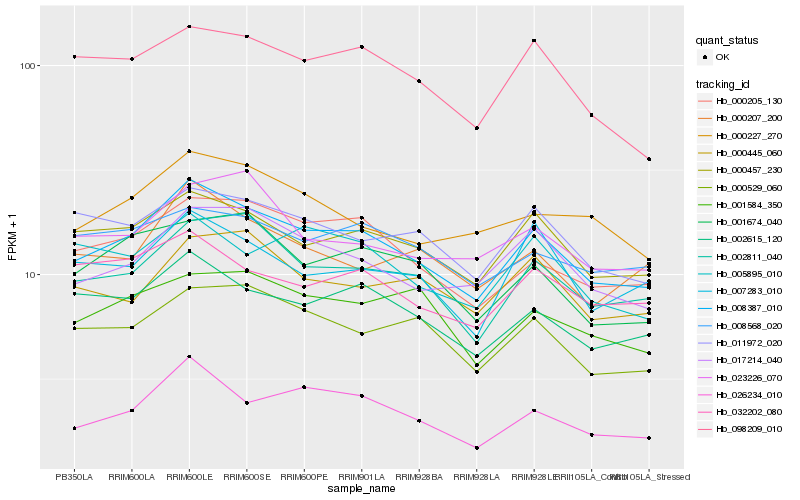
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008387_010 |
0.0 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 2 |
Hb_000457_230 |
0.0659501797 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002811_040 |
0.0675939851 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 4 |
Hb_000205_130 |
0.0681027137 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_002615_120 |
0.0705798648 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_017214_040 |
0.071364182 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_026234_010 |
0.0765095259 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_000529_060 |
0.0791618696 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 9 |
Hb_098209_010 |
0.0805257173 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 10 |
Hb_001674_040 |
0.082927938 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 11 |
Hb_011972_020 |
0.0833526515 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_000227_270 |
0.0845313889 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_032202_080 |
0.0861825746 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000207_200 |
0.0875053662 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001584_350 |
0.0887227751 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 16 |
Hb_008568_020 |
0.0887680537 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 17 |
Hb_000445_060 |
0.0891243705 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 18 |
Hb_007283_010 |
0.0895704813 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 19 |
Hb_023226_070 |
0.089793191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005895_010 |
0.0908509577 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |