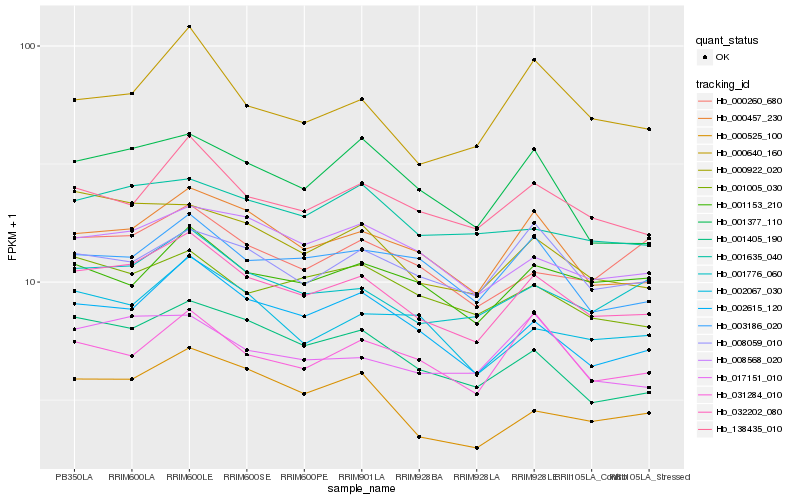
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032202_080 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000457_230 |
0.0554879255 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002615_120 |
0.0578358278 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001776_060 |
0.0588721495 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_138435_010 |
0.062731554 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 6 |
Hb_008568_020 |
0.0668294682 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 7 |
Hb_001405_190 |
0.0699383815 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_003186_020 |
0.0709216791 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000922_020 |
0.071266909 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_002067_030 |
0.0714669127 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001153_210 |
0.0731944967 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000525_100 |
0.073298201 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 13 |
Hb_001005_030 |
0.0734439455 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000640_160 |
0.0737021964 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 15 |
Hb_001635_040 |
0.0745014785 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_031284_010 |
0.0753096128 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 17 |
Hb_017151_010 |
0.0756657483 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 18 |
Hb_000260_680 |
0.0763070542 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 19 |
Hb_008059_010 |
0.0769158169 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001377_110 |
0.0770424518 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |