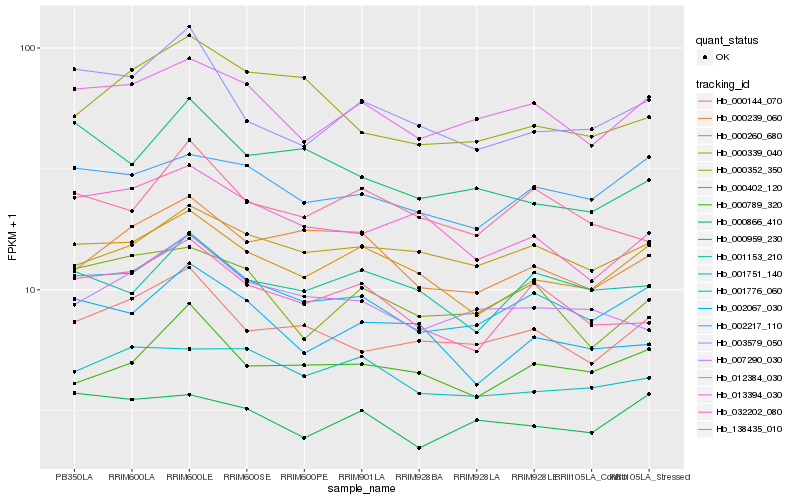
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_060 |
0.0 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 2 |
Hb_012384_030 |
0.0489601359 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 3 |
Hb_000260_680 |
0.0564902442 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 4 |
Hb_000144_070 |
0.0584197774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_032202_080 |
0.0588721495 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002217_110 |
0.0642111343 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 7 |
Hb_007290_030 |
0.0716527085 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 8 |
Hb_001153_210 |
0.0740338459 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000402_120 |
0.0743333437 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 10 |
Hb_000239_060 |
0.0747393875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003579_050 |
0.0749730891 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 12 |
Hb_000339_040 |
0.0760320824 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_000789_320 |
0.0782748297 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_013394_030 |
0.078301688 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 15 |
Hb_001751_140 |
0.0786202144 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000866_410 |
0.0787586431 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 17 |
Hb_000959_230 |
0.079440731 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 18 |
Hb_000352_350 |
0.0809839887 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 19 |
Hb_138435_010 |
0.0810046623 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 20 |
Hb_002067_030 |
0.0811771053 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |