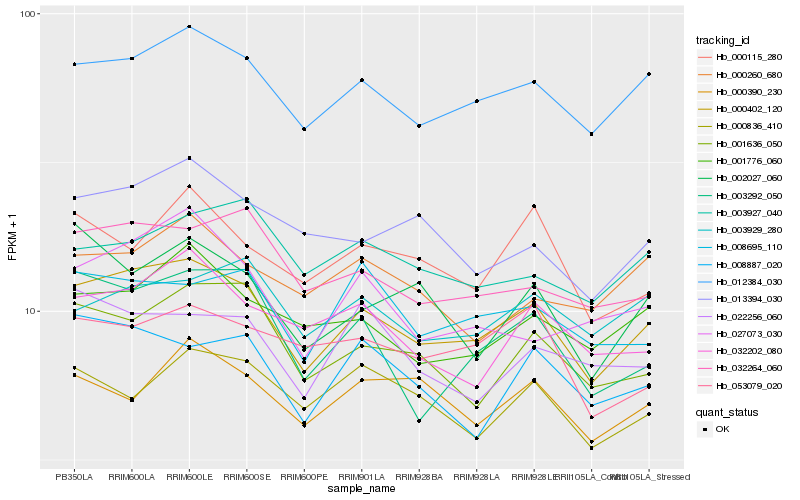
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 2 |
Hb_012384_030 |
0.0476716506 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 3 |
Hb_032264_060 |
0.0724694681 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 4 |
Hb_001776_060 |
0.0743333437 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_001636_050 |
0.0754501518 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 6 |
Hb_013394_030 |
0.0796430211 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 7 |
Hb_032202_080 |
0.0811240597 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_008695_110 |
0.0815864394 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008887_020 |
0.0821430439 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 10 |
Hb_003929_280 |
0.0824965514 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 11 |
Hb_003292_050 |
0.0830731663 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 12 |
Hb_003927_040 |
0.0849403463 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 13 |
Hb_000390_230 |
0.0851350895 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 14 |
Hb_000115_280 |
0.0853629157 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 15 |
Hb_027073_030 |
0.0857540691 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 16 |
Hb_002027_060 |
0.0871044204 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 17 |
Hb_000836_410 |
0.087223358 |
- |
- |
sec10, putative [Ricinus communis] |
| 18 |
Hb_000260_680 |
0.0873837903 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 19 |
Hb_053079_020 |
0.0884562649 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 20 |
Hb_022256_060 |
0.0885719734 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |