| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
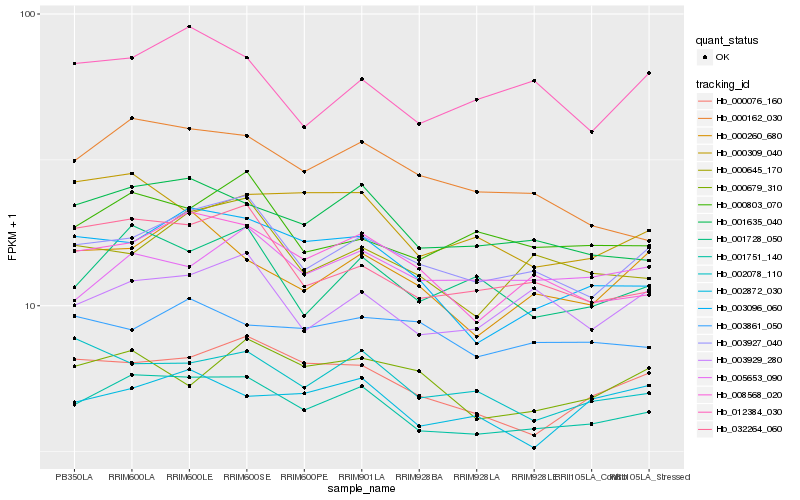
Hb_001751_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001635_040 |
0.0475523177 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_003927_040 |
0.0538427567 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 4 |
Hb_008568_020 |
0.0579531418 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 5 |
Hb_000076_160 |
0.0605568827 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 6 |
Hb_001728_050 |
0.0612092655 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 7 |
Hb_000803_070 |
0.0632721186 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 8 |
Hb_003929_280 |
0.0640679855 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 9 |
Hb_002872_030 |
0.066548744 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 10 |
Hb_032264_060 |
0.0672549048 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 11 |
Hb_002078_110 |
0.0676775516 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_005653_090 |
0.0680037479 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 13 |
Hb_000309_040 |
0.0703568 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 14 |
Hb_000260_680 |
0.0707067829 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 15 |
Hb_000645_170 |
0.0710932048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000679_310 |
0.0714924888 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003096_060 |
0.0715864067 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 18 |
Hb_003861_050 |
0.0727761477 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 19 |
Hb_000162_030 |
0.0727826123 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_012384_030 |
0.0730443071 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |