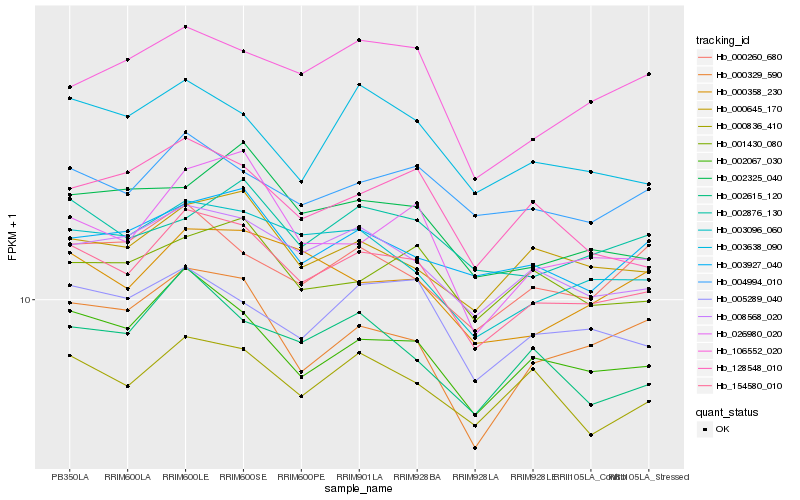
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_154580_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 2 |
Hb_002067_030 |
0.0515479735 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_008568_020 |
0.0526638789 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 4 |
Hb_003096_060 |
0.0563162503 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 5 |
Hb_000645_170 |
0.0626560405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005289_040 |
0.0645288262 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 7 |
Hb_026980_020 |
0.065073407 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 8 |
Hb_000836_410 |
0.0663071774 |
- |
- |
sec10, putative [Ricinus communis] |
| 9 |
Hb_004994_010 |
0.0671992918 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_000358_230 |
0.0680318036 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 11 |
Hb_003638_090 |
0.0705386966 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 12 |
Hb_001430_080 |
0.0707695385 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 13 |
Hb_003927_040 |
0.0715448951 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 14 |
Hb_106552_020 |
0.0727248075 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002325_040 |
0.0728999665 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002615_120 |
0.0729494041 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000260_680 |
0.0734059975 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 18 |
Hb_128548_010 |
0.073583112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002876_130 |
0.0745225709 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 20 |
Hb_000329_590 |
0.0746759223 |
- |
- |
catalytic, putative [Ricinus communis] |