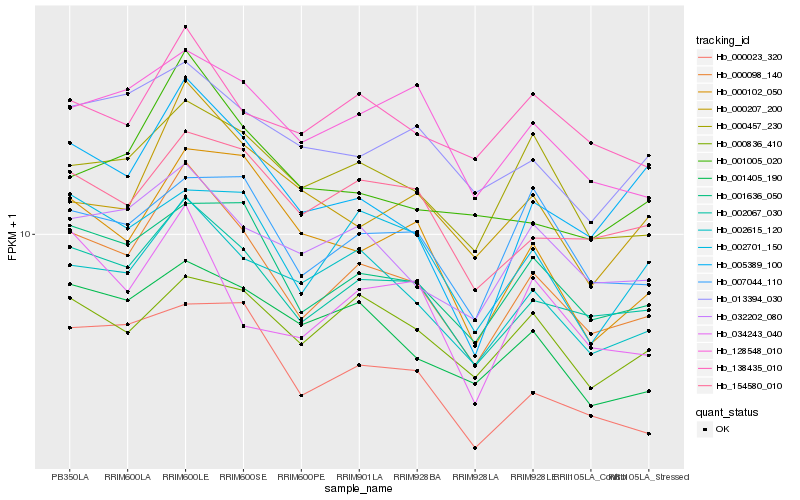
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 2 |
Hb_002067_030 |
0.049047143 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002615_120 |
0.0677101874 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001636_050 |
0.0704607509 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 5 |
Hb_032202_080 |
0.0786994489 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000457_230 |
0.0827827748 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000023_320 |
0.0844297261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001405_190 |
0.0853748755 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_154580_010 |
0.0860094964 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 10 |
Hb_005389_100 |
0.0877627616 |
- |
- |
PREDICTED: methyltransferase-like protein 5 isoform X2 [Jatropha curcas] |
| 11 |
Hb_128548_010 |
0.0882354721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002701_150 |
0.0889860115 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000102_050 |
0.0897059729 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001005_020 |
0.0904310224 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 15 |
Hb_034243_040 |
0.0915704863 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 16 |
Hb_013394_030 |
0.0917618796 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 17 |
Hb_007044_110 |
0.0917705051 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000207_200 |
0.092840397 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_138435_010 |
0.0931974792 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 20 |
Hb_000836_410 |
0.0932259278 |
- |
- |
sec10, putative [Ricinus communis] |