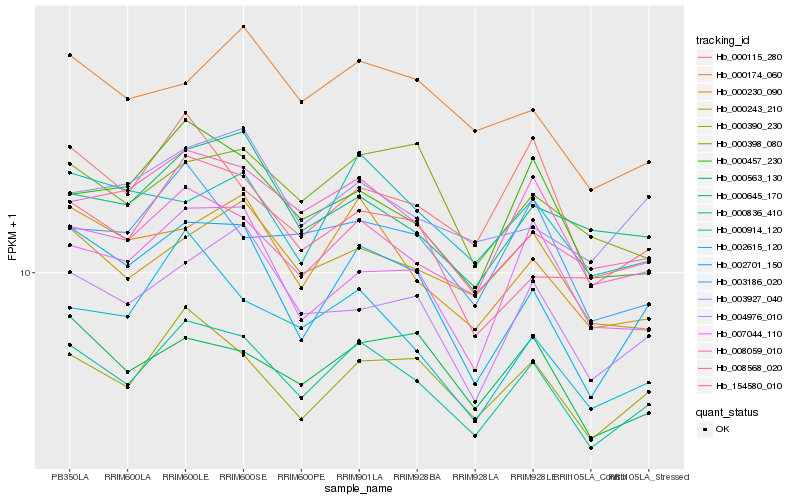
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_410 |
0.0 |
- |
- |
sec10, putative [Ricinus communis] |
| 2 |
Hb_000390_230 |
0.0473026771 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 3 |
Hb_008568_020 |
0.0548711718 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 4 |
Hb_000230_090 |
0.0590711742 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002701_150 |
0.0592005671 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000457_230 |
0.0614129881 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000243_210 |
0.0618067894 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000563_130 |
0.0632746153 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 9 |
Hb_008059_010 |
0.0651291222 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004976_010 |
0.0654407293 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 11 |
Hb_000914_120 |
0.0656446267 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 12 |
Hb_154580_010 |
0.0663071774 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 13 |
Hb_000645_170 |
0.0664374913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003186_020 |
0.0664702447 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002615_120 |
0.0669249419 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000115_280 |
0.0673253834 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_007044_110 |
0.0684536284 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000398_080 |
0.0703171352 |
- |
- |
tip120, putative [Ricinus communis] |
| 19 |
Hb_000174_060 |
0.070462869 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_003927_040 |
0.0705229354 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |