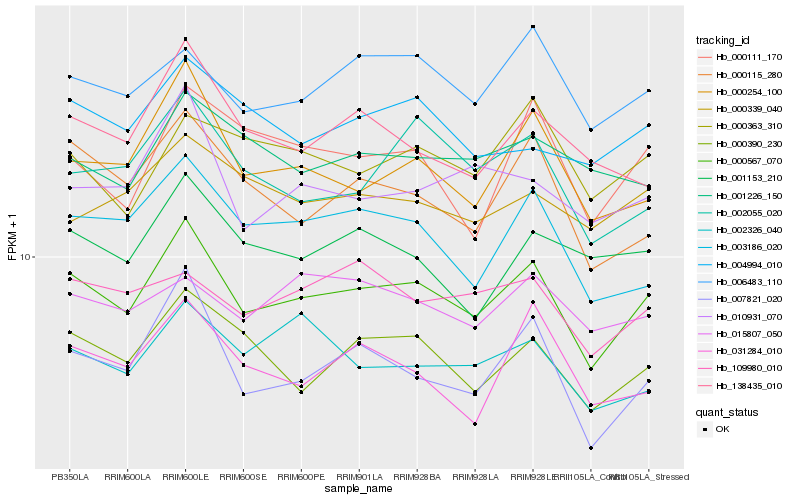
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_070 |
0.0 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 2 |
Hb_007821_020 |
0.0591766282 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000390_230 |
0.0612218945 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 4 |
Hb_000254_100 |
0.0629593893 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 5 |
Hb_003186_020 |
0.0671265458 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006483_110 |
0.0676030229 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 7 |
Hb_000115_280 |
0.0689344332 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 8 |
Hb_002055_020 |
0.0728871605 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 9 |
Hb_031284_010 |
0.0747715064 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 10 |
Hb_002326_040 |
0.0750737328 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 11 |
Hb_138435_010 |
0.0752286778 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 12 |
Hb_004994_010 |
0.0754312836 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_010931_070 |
0.0759550254 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000111_170 |
0.0760395967 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001153_210 |
0.0770381169 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000363_310 |
0.0770499035 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 17 |
Hb_109980_010 |
0.0777027013 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 18 |
Hb_015807_050 |
0.0790295868 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 19 |
Hb_000339_040 |
0.0796672405 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_001226_150 |
0.0798770217 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |