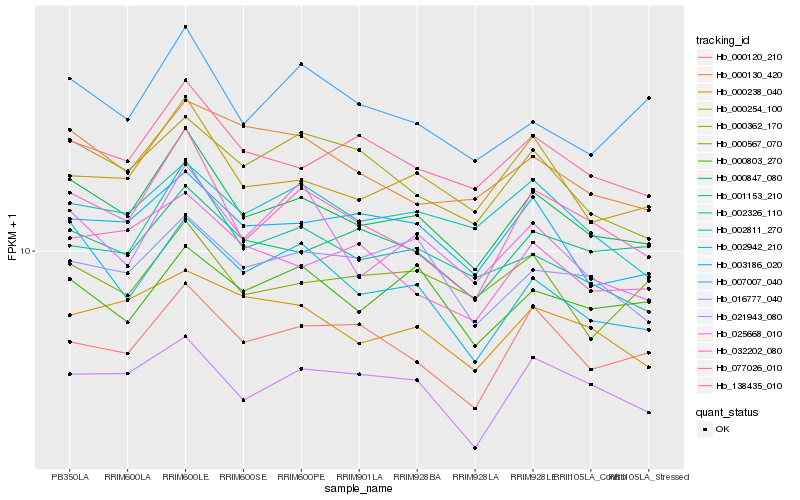
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000847_080 |
0.0 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 2 |
Hb_077026_010 |
0.0566274939 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 3 |
Hb_000254_100 |
0.058963797 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 4 |
Hb_138435_010 |
0.0623743648 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 5 |
Hb_002326_110 |
0.0641474223 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 6 |
Hb_001153_210 |
0.0711958041 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003186_020 |
0.0720697685 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 8 |
Hb_021943_080 |
0.0744626638 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002942_210 |
0.0749071053 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 10 |
Hb_007007_040 |
0.076612903 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 11 |
Hb_000120_210 |
0.0772615504 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 12 |
Hb_032202_080 |
0.0774003228 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000238_040 |
0.0777688929 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 14 |
Hb_025668_010 |
0.0779409973 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000130_420 |
0.0784227858 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 16 |
Hb_000362_170 |
0.0789806254 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 17 |
Hb_002811_270 |
0.0796184386 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 18 |
Hb_016777_040 |
0.080709931 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 19 |
Hb_000803_270 |
0.0810132923 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 20 |
Hb_000567_070 |
0.0814117389 |
- |
- |
autophagy protein, putative [Ricinus communis] |