| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
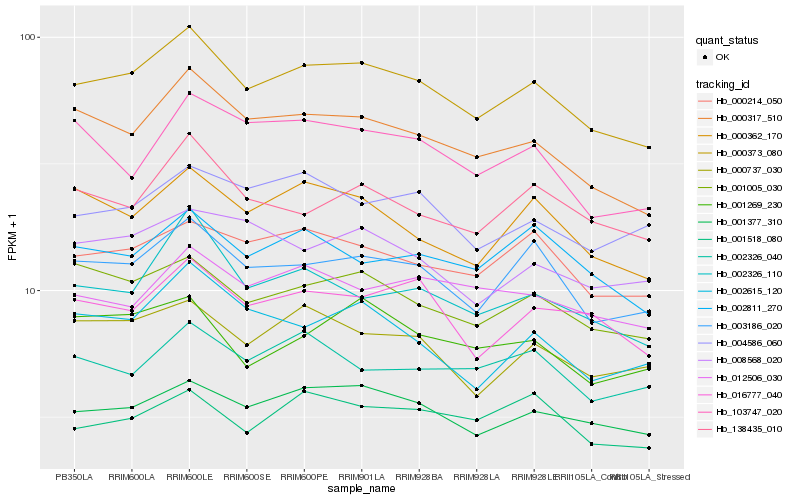
Hb_000373_080 |
0.0 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 2 |
Hb_003186_020 |
0.0491074424 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000317_510 |
0.0498676271 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 4 |
Hb_001377_310 |
0.0514778307 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 5 |
Hb_001005_030 |
0.0558735336 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000214_050 |
0.0584008562 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000362_170 |
0.0624630758 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 8 |
Hb_016777_040 |
0.0634806757 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 9 |
Hb_000737_030 |
0.0653698156 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002811_270 |
0.0655943769 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 11 |
Hb_002326_110 |
0.0659912967 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 12 |
Hb_138435_010 |
0.0664274549 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 13 |
Hb_012506_030 |
0.0702977492 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 14 |
Hb_008568_020 |
0.0714718944 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 15 |
Hb_001269_230 |
0.0715044506 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 16 |
Hb_001518_080 |
0.0717243397 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 17 |
Hb_004586_060 |
0.0719543049 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 18 |
Hb_002615_120 |
0.0724028095 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_103747_020 |
0.0738928227 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 20 |
Hb_002326_040 |
0.0742567886 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |