| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
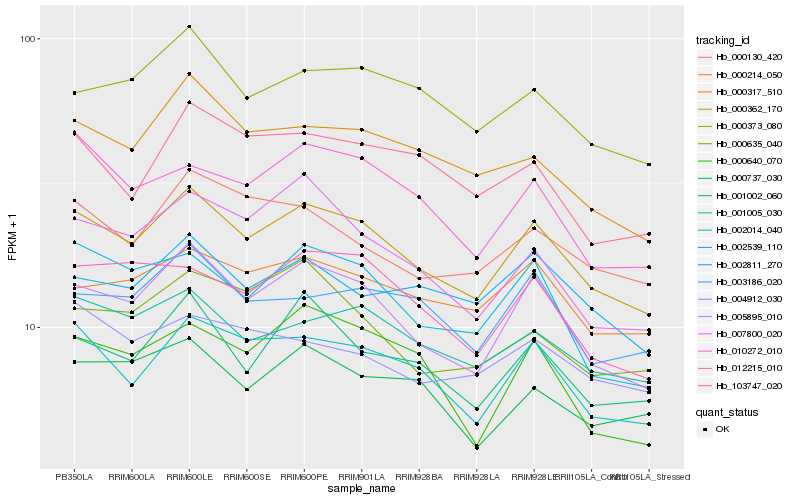
Hb_000362_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 2 |
Hb_005895_010 |
0.0355794329 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002014_040 |
0.0481211951 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 4 |
Hb_001005_030 |
0.0555085442 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_001002_060 |
0.0592756471 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 6 |
Hb_000214_050 |
0.0619247497 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000373_080 |
0.0624630758 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 8 |
Hb_002539_110 |
0.0627563123 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 9 |
Hb_000317_510 |
0.0629159925 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 10 |
Hb_012215_010 |
0.0630213235 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 11 |
Hb_010272_010 |
0.0631318589 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 12 |
Hb_003186_020 |
0.0642831772 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000737_030 |
0.0663573837 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000130_420 |
0.066811016 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_007800_020 |
0.0685829388 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002811_270 |
0.0686978707 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 17 |
Hb_000640_070 |
0.0702787121 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 18 |
Hb_103747_020 |
0.0703344209 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 19 |
Hb_004912_030 |
0.0723526772 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 20 |
Hb_000635_040 |
0.072511618 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |