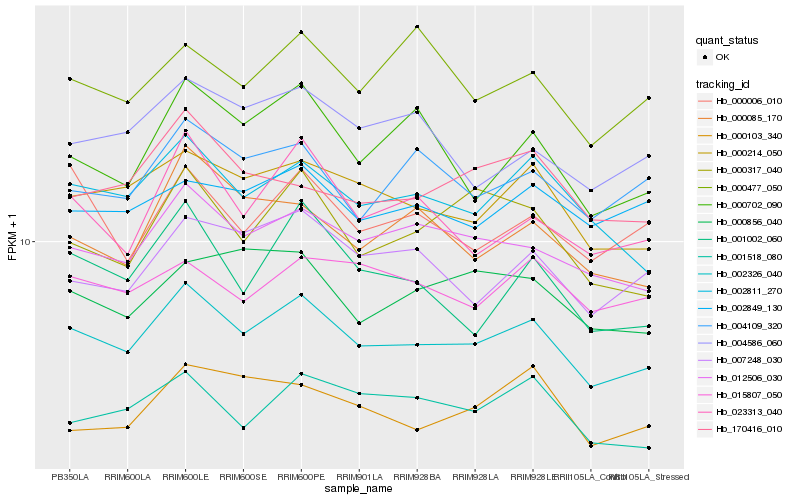
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002326_040 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 2 |
Hb_000214_050 |
0.0500983546 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012506_030 |
0.0507576627 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 4 |
Hb_002811_270 |
0.053949241 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 5 |
Hb_001518_080 |
0.0647980236 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 6 |
Hb_007248_030 |
0.0656523121 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 7 |
Hb_023313_040 |
0.0657729775 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004109_320 |
0.0670333694 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 9 |
Hb_002849_130 |
0.0674936369 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 10 |
Hb_000317_040 |
0.0686739161 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_015807_050 |
0.068875564 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 12 |
Hb_170416_010 |
0.0691986374 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 13 |
Hb_000006_010 |
0.0693415727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000856_040 |
0.0693621999 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 15 |
Hb_000085_170 |
0.0700721886 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001002_060 |
0.0704266039 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 17 |
Hb_004586_060 |
0.0704700262 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 18 |
Hb_000702_090 |
0.0712621066 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 19 |
Hb_000103_340 |
0.0725143454 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 20 |
Hb_000477_050 |
0.073075586 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |