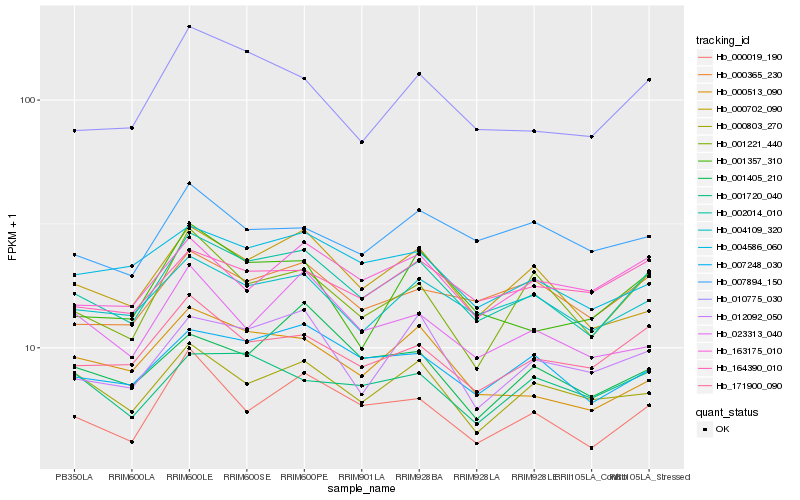
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002014_010 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_000019_190 |
0.0529711941 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 3 |
Hb_007248_030 |
0.0562058002 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 4 |
Hb_004109_320 |
0.0565133033 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 5 |
Hb_171900_090 |
0.06355348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000702_090 |
0.0661244097 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_000365_230 |
0.0666238059 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 8 |
Hb_004586_060 |
0.0668537074 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 9 |
Hb_012092_050 |
0.0686728324 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000513_090 |
0.0690749072 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 11 |
Hb_000803_270 |
0.0692425407 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 12 |
Hb_001221_440 |
0.0692508141 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010775_030 |
0.0695024777 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 14 |
Hb_164390_010 |
0.0710995689 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 15 |
Hb_163175_010 |
0.0712818789 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 16 |
Hb_023313_040 |
0.0759034202 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001720_040 |
0.076980284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001405_210 |
0.0770593651 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 19 |
Hb_007894_150 |
0.0774208664 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 20 |
Hb_001357_310 |
0.0781049375 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |