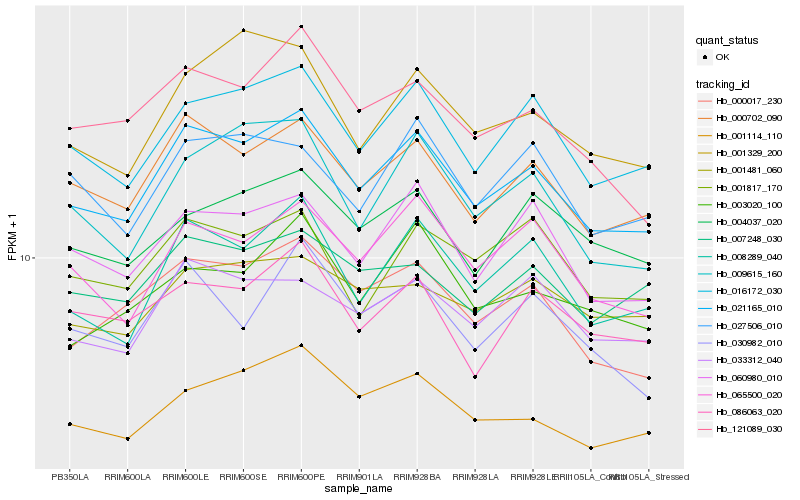
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021165_010 |
0.0 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000702_090 |
0.0414976166 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 3 |
Hb_016172_030 |
0.0496392605 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 4 |
Hb_008289_040 |
0.0539433119 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 5 |
Hb_000017_230 |
0.0593026016 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 6 |
Hb_060980_010 |
0.061048359 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_001114_110 |
0.0618861698 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 8 |
Hb_065500_020 |
0.0619335925 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 9 |
Hb_033312_040 |
0.0626497963 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_121089_030 |
0.0666907463 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 11 |
Hb_009615_160 |
0.0672689598 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 12 |
Hb_007248_030 |
0.0702707519 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 13 |
Hb_001329_200 |
0.0706127652 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 14 |
Hb_004037_020 |
0.0715068529 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 15 |
Hb_027506_010 |
0.0729110993 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 16 |
Hb_086063_020 |
0.073233359 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 17 |
Hb_003020_100 |
0.0734619963 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 18 |
Hb_001817_170 |
0.0738957432 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 19 |
Hb_001481_060 |
0.0743812029 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_030982_010 |
0.0743988348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |