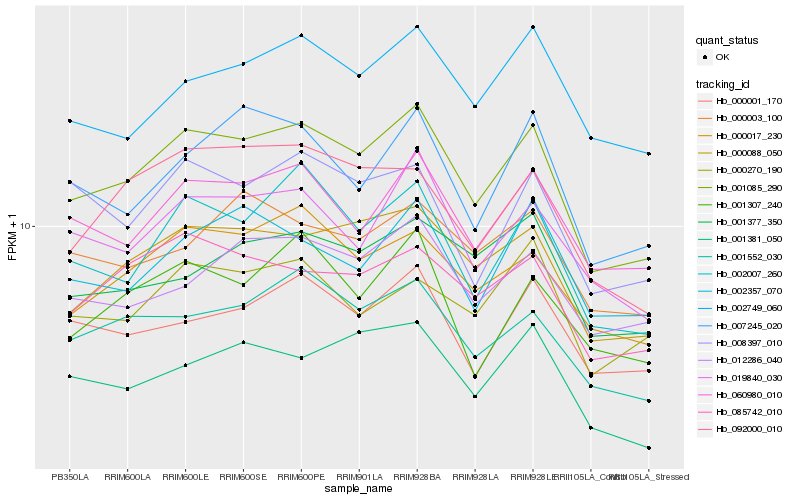
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_290 |
0.0 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 2 |
Hb_000088_050 |
0.0581712843 |
- |
- |
PREDICTED: threonylcarbamoyladenosine tRNA methylthiotransferase [Jatropha curcas] |
| 3 |
Hb_000017_230 |
0.0645264298 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 4 |
Hb_002007_260 |
0.0656987374 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 5 |
Hb_001552_030 |
0.0663323908 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_060980_010 |
0.0673492945 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_085742_010 |
0.0714364727 |
- |
- |
hypothetical protein POPTR_0005s05170g [Populus trichocarpa] |
| 8 |
Hb_019840_030 |
0.0714505636 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000001_170 |
0.0766006927 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_002749_060 |
0.0769189228 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 11 |
Hb_012286_040 |
0.0776873443 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 12 |
Hb_092000_010 |
0.0785428149 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 13 |
Hb_007245_020 |
0.0790613842 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 14 |
Hb_002357_070 |
0.0795536922 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 15 |
Hb_001381_050 |
0.0800490974 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 16 |
Hb_008397_010 |
0.0825304642 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001307_240 |
0.0832834851 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001377_350 |
0.0833593397 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 19 |
Hb_000003_100 |
0.0839453579 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 20 |
Hb_000270_190 |
0.0839606727 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |