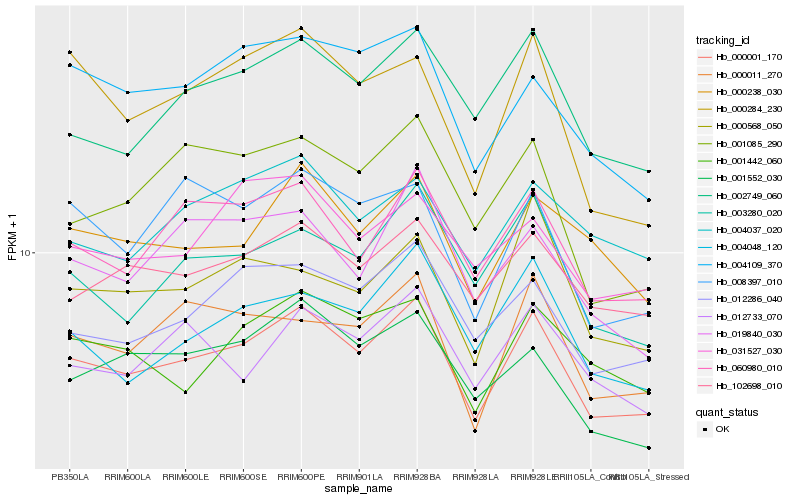
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_170 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_060980_010 |
0.0607100267 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_012286_040 |
0.0649386031 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 4 |
Hb_102698_010 |
0.0670603579 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002749_060 |
0.0683012233 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 6 |
Hb_000011_270 |
0.0703231317 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 7 |
Hb_000238_030 |
0.0703543866 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_003280_020 |
0.0712921776 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001552_030 |
0.0724528822 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004048_120 |
0.0727996866 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 11 |
Hb_004109_370 |
0.0730522334 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_001442_060 |
0.0738516061 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001085_290 |
0.0766006927 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 14 |
Hb_012733_070 |
0.0767334936 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 15 |
Hb_019840_030 |
0.0768035445 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 16 |
Hb_008397_010 |
0.0774490186 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000568_050 |
0.0783136149 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000284_230 |
0.0783939644 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 19 |
Hb_004037_020 |
0.0786935134 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 20 |
Hb_031527_030 |
0.0790582872 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |