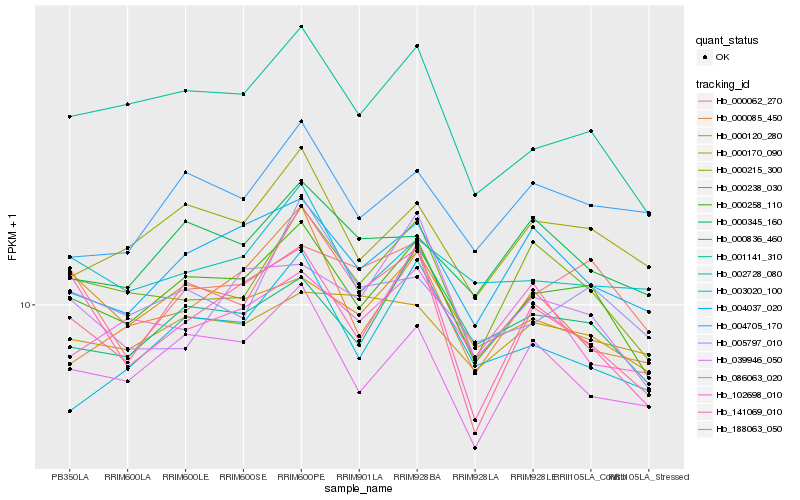
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_110 |
0.0 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001141_310 |
0.0538668065 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 3 |
Hb_000836_460 |
0.0604621288 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 4 |
Hb_086063_020 |
0.0632853528 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000238_030 |
0.0637221542 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000215_300 |
0.0637330559 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_004037_020 |
0.0657380463 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000062_270 |
0.0726406036 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000345_160 |
0.0759971406 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 10 |
Hb_141069_010 |
0.0762198484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005797_010 |
0.0778286646 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_000170_090 |
0.0783012959 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 13 |
Hb_188063_050 |
0.0783144119 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 14 |
Hb_000120_280 |
0.0792159553 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 15 |
Hb_002728_080 |
0.0811043672 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_004705_170 |
0.0833045079 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 17 |
Hb_000085_450 |
0.0844759052 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 18 |
Hb_039946_050 |
0.0857911645 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_003020_100 |
0.0864532812 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 20 |
Hb_102698_010 |
0.0868641761 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |