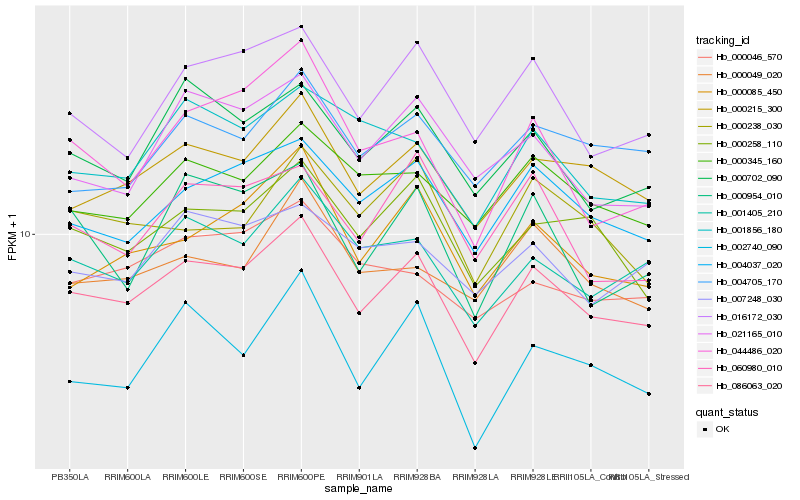
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086063_020 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000215_300 |
0.0562697478 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_004037_020 |
0.0578406352 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001405_210 |
0.0626085878 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 5 |
Hb_000258_110 |
0.0632853528 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000702_090 |
0.0643974854 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_016172_030 |
0.0649938356 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 8 |
Hb_000085_450 |
0.0664841424 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 9 |
Hb_000345_160 |
0.0683370933 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 10 |
Hb_021165_010 |
0.073233359 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_060980_010 |
0.0734023274 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 12 |
Hb_004705_170 |
0.074579236 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 13 |
Hb_000954_010 |
0.0749898894 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 14 |
Hb_001856_180 |
0.0754658685 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 15 |
Hb_000049_020 |
0.0758536062 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 16 |
Hb_007248_030 |
0.0762598457 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 17 |
Hb_000046_570 |
0.0765505791 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 18 |
Hb_044486_020 |
0.0770487834 |
- |
- |
CASTOR protein [Glycine max] |
| 19 |
Hb_000238_030 |
0.0781287553 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_002740_090 |
0.0783226511 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |