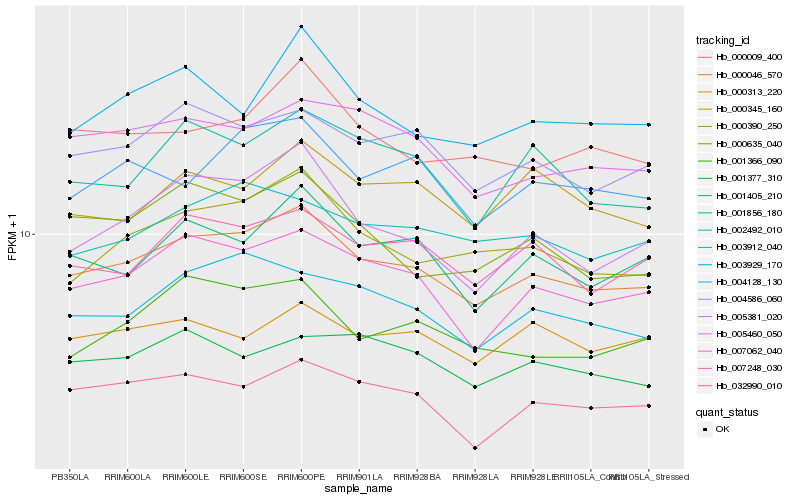
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_570 |
0.0 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 2 |
Hb_007062_040 |
0.0497235565 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 3 |
Hb_004586_060 |
0.0539750326 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 4 |
Hb_007248_030 |
0.0590068272 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 5 |
Hb_000390_250 |
0.05902177 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 6 |
Hb_001405_210 |
0.059083803 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 7 |
Hb_003912_040 |
0.0605620312 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 8 |
Hb_005381_020 |
0.060929284 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 9 |
Hb_005460_050 |
0.0628106406 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 10 |
Hb_004128_130 |
0.0651837999 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002492_010 |
0.0660014735 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 12 |
Hb_000009_400 |
0.0663798263 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 13 |
Hb_001856_180 |
0.0664133709 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 14 |
Hb_032990_010 |
0.0674936793 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 15 |
Hb_003929_170 |
0.0683634119 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 16 |
Hb_001366_090 |
0.0695478463 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 17 |
Hb_000635_040 |
0.0706785403 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 18 |
Hb_000345_160 |
0.0732220078 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 19 |
Hb_000313_220 |
0.073591266 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001377_310 |
0.0741746022 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |