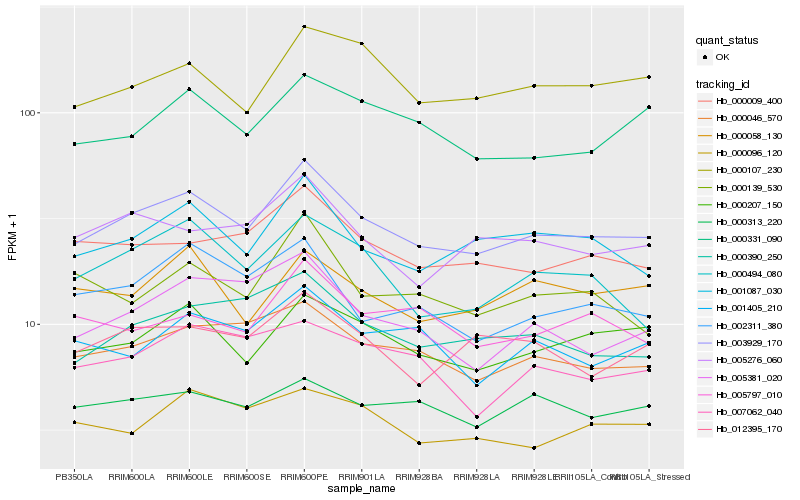
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_170 |
0.0 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 2 |
Hb_001087_030 |
0.0669483952 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 3 |
Hb_000046_570 |
0.0683634119 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 4 |
Hb_000207_150 |
0.0696532228 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 5 |
Hb_000009_400 |
0.0708545902 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 6 |
Hb_000390_250 |
0.075806169 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 7 |
Hb_000107_230 |
0.0771272171 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_005276_060 |
0.078317867 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_012395_170 |
0.0797345463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000331_090 |
0.0807808978 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 11 |
Hb_002311_380 |
0.081478823 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 12 |
Hb_005381_020 |
0.0823145167 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 13 |
Hb_001405_210 |
0.0825423518 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 14 |
Hb_000494_080 |
0.083575529 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000096_120 |
0.0849360284 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 16 |
Hb_000058_130 |
0.0855652072 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 17 |
Hb_005797_010 |
0.0861624392 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_000313_220 |
0.0864227491 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007062_040 |
0.0867849683 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 20 |
Hb_000139_530 |
0.0868367231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |